|
Puromycin
Puromycin is an antibiotic protein synthesis inhibitor which causes premature chain termination during translation. Inhibition of translation Puromycin is an aminonucleoside antibiotic, derived from the '' Streptomyces alboniger'' bacterium, that causes premature chain termination during translation taking place in the ribosome. Part of the molecule resembles the 3' end of the aminoacylated tRNA. It enters the A site and transfers to the growing chain, causing the formation of a puromycylated nascent chain and premature chain release. The exact mechanism of action is unknown at this time but the 3' position contains an amide linkage instead of the normal ester linkage of tRNA. That makes the molecule much more resistant to hydrolysis and stops the ribosome. Puromycin is selective for either prokaryotes or eukaryotes. Also of note, puromycin is critical in mRNA display. In this reaction, a puromycin molecule is chemically attached to the end of an mRNA template, which is then ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA Display
mRNA display is a display technique used for ''in vitro'' protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step (affinity chromatography). The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest. Puromycin is an analogue of the 3’ end of a tyrosyl-tRNA with a part of its structure mimics a molecule of adenosine Adenosine ( symbol A) is an organic compound that occurs widely in nature in the form of diverse derivatives. The molecule consists of an adenine attached to a ribose via a β-N9-glycosidic bond. Adenosine is one of the four nucleoside building ..., and t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Synthesis Inhibitor
A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins. While a broad interpretation of this definition could be used to describe nearly any compound depending on concentration, in practice, it usually refers to compounds that act at the molecular level on translational machinery (either the ribosome itself or the translation factor), taking advantages of the major differences between prokaryotic and eukaryotic ribosome structures. Mechanism In general, protein synthesis inhibitors work at different stages of bacterial mRNA translation into proteins, like initiation, elongation (including aminoacyl tRNA entry, proofreading, peptidyl transfer, and bacterial translocation) and termination: Earlier stages * Rifamycin inhibits bacterial DNA transcription into mRNA by inhibiting DNA-dependent RNA polymerase by binding its beta-subunit. * alpha-Amanitin is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Streptomyces Alboniger
''Streptomyces alboniger'' is a bacterium species in the genus '' Streptomyces''. ''Streptomyces alboniger'' produces puromycin Puromycin is an antibiotic protein synthesis inhibitor which causes premature chain termination during translation. Inhibition of translation Puromycin is an aminonucleoside antibiotic, derived from the '' Streptomyces alboniger'' bacterium ..., a type of antibiotic. See also * List of ''Streptomyces'' species References External links Type strain of ''Streptomyces alboniger'' at Bac''Dive'' - the Bacterial Diversity Metadatabase alboniger Bacteria described in 1952 {{Streptomyces-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free Base
Free base (freebase, free-base) is the conjugate base (deprotonated) form of an amine, as opposed to its conjugate acid (protonated) form. The amine is often an alkaloid, such as nicotine, cocaine, morphine, and ephedrine, or derivatives thereof. Freebasing is a more efficient method of self-administering alkaloids via the smoking route. Properties Some alkaloids are more stable as ionic salts than as free base. The salts usually exhibit greater water solubility. Common counterions include chloride, bromide, sulfate, phosphate, nitrate, acetate, oxalate, citrate, and tartrate. Ammonium salts formed from the acid-base reaction with hydrochloric acid are known as hydrochlorides. For example, compare the free base hydroxylamine (NH2OH) with the salt hydroxylamine hydrochloride (NH3OH+ Cl−). Freebasing Cocaine hydrochloride ("powder cocaine") cannot be smoked as it decomposes at the high temperatures produced by smoking. Free base cocaine, on the other hand, has a melting poin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosides
Nucleosides are glycosylamines that can be thought of as nucleotides without a phosphate group. A nucleoside consists simply of a nucleobase (also termed a nitrogenous base) and a five-carbon sugar (ribose or 2'-deoxyribose) whereas a nucleotide is composed of a nucleobase, a five-carbon sugar, and one or more phosphate groups. In a nucleoside, the anomeric carbon is linked through a glycosidic bond to the N9 of a purine or the N1 of a pyrimidine. Nucleotides are the molecular building-blocks of DNA and RNA. List of nucleosides and corresponding nucleobases The reason for 2 symbols, shorter and longer, is that the shorter ones are better for contexts where explicit disambiguation is superfluous (because context disambiguates) and the longer ones are for contexts where explicit disambiguation is judged to be needed or wise. For example, when discussing long nucleobase sequences in genomes, the CATG symbol system is much preferable to the Cyt-Ade-Thy-Gua symbol system (see '' N ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Synthesis Inhibitor Antibiotics
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
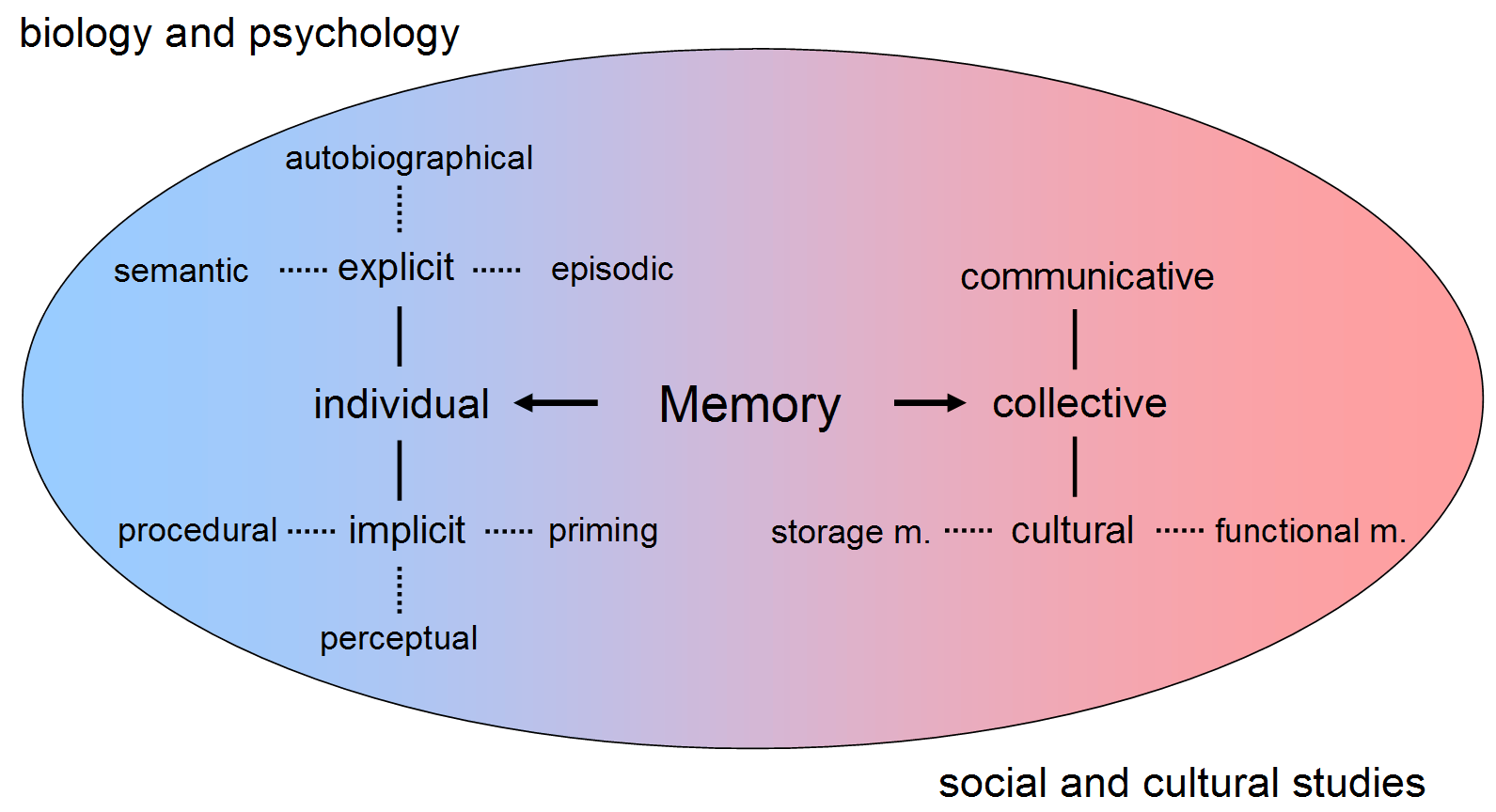
Memory
Memory is the faculty of the mind by which data or information is encoded, stored, and retrieved when needed. It is the retention of information over time for the purpose of influencing future action. If past events could not be remembered, it would be impossible for language, relationships, or personal identity to develop. Memory loss is usually described as forgetfulness or amnesia. Memory is often understood as an informational processing system with explicit and implicit functioning that is made up of a sensory processor, short-term (or working) memory, and long-term memory. This can be related to the neuron. The sensory processor allows information from the outside world to be sensed in the form of chemical and physical stimuli and attended to various levels of focus and intent. Working memory serves as an encoding and retrieval processor. Information in the form of stimuli is encoded in accordance with explicit or implicit functions by the working memory processor. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synaptic Plasticity
In neuroscience, synaptic plasticity is the ability of synapses to strengthen or weaken over time, in response to increases or decreases in their activity. Since memories are postulated to be represented by vastly interconnected neural circuits in the brain, synaptic plasticity is one of the important neurochemical foundations of learning and memory (''see Hebbian theory''). Plastic change often results from the alteration of the number of neurotransmitter receptors located on a synapse. There are several underlying mechanisms that cooperate to achieve synaptic plasticity, including changes in the quantity of neurotransmitters released into a synapse and changes in how effectively cells respond to those neurotransmitters. Synaptic plasticity in both excitatory and inhibitory synapses has been found to be dependent upon postsynaptic calcium release. Historical discoveries In 1973, Terje Lømo and Tim Bliss first described the now widely studied phenomenon of long-term pote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metallopeptidase
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis. Most metalloproteases require zinc, but some use cobalt. The metal ion is coordinated to the protein via three ligands. The ligands coordinating the metal ion can vary with histidine, glutamate, aspartate, lysine, and arginine. The fourth coordination position is taken up by a labile water molecule. Treatment with chelating agents such as EDTA leads to complete inactivation. EDTA is a metal chelator that removes zinc, which is essential for activity. They are also inhibited by the chelator orthophenanthroline. Classification There are two subgroups of metalloproteinases: * Exopeptidases, metalloexopeptidases ( EC number: 3.4.17). * Endopeptidases, metalloendopeptidases (3.4.24). Well known metalloendopeptidases include ADAM pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Protease
Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like. Classification The MEROPS protease classification system counts 16 superfamilies (as of 2013) each containing many families. Each superfamily uses the catalytic triad or dyad in a different protein fold and so represent convergent evolution of the catalytic mechanism. The majority belong to the S1 family of the PA clan (superfamily) of proteases. For superfamilies, P: superfamily, containing a mixture of nucleophile class families, S: purely serine proteases. superfamily. Within each superfamily, families are designated by their catalytic nucleophile, (S: serine proteases). Substrate specificity Serine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
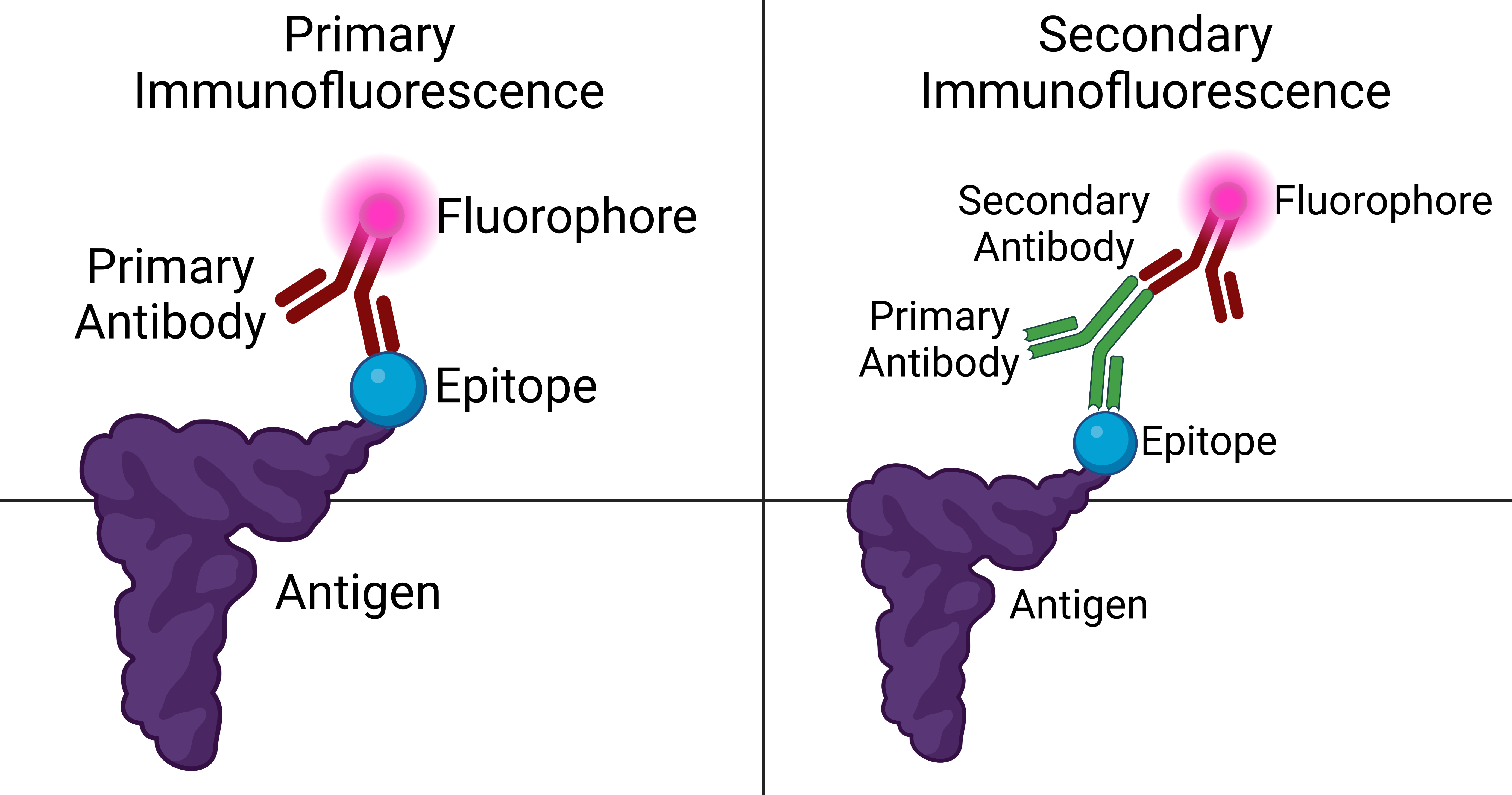
Immunofluorescence
Immunofluorescence is a technique used for light microscopy with a fluorescence microscope and is used primarily on microbiological samples. This technique uses the specificity of antibodies to their antigen to target fluorescent dyes to specific biomolecule targets within a cell, and therefore allows visualization of the distribution of the target molecule through the sample. The specific region an antibody recognizes on an antigen is called an epitope. There have been efforts in epitope mapping since many antibodies can bind the same epitope and levels of binding between antibodies that recognize the same epitope can vary. Additionally, the binding of the fluorophore to the antibody itself cannot interfere with the immunological specificity of the antibody or the binding capacity of its antigen. Immunofluorescence is a widely used example of immunostaining (using antibodies to stain proteins) and is a specific example of immunohistochemistry (the use of the antibody-antigen rel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |