|
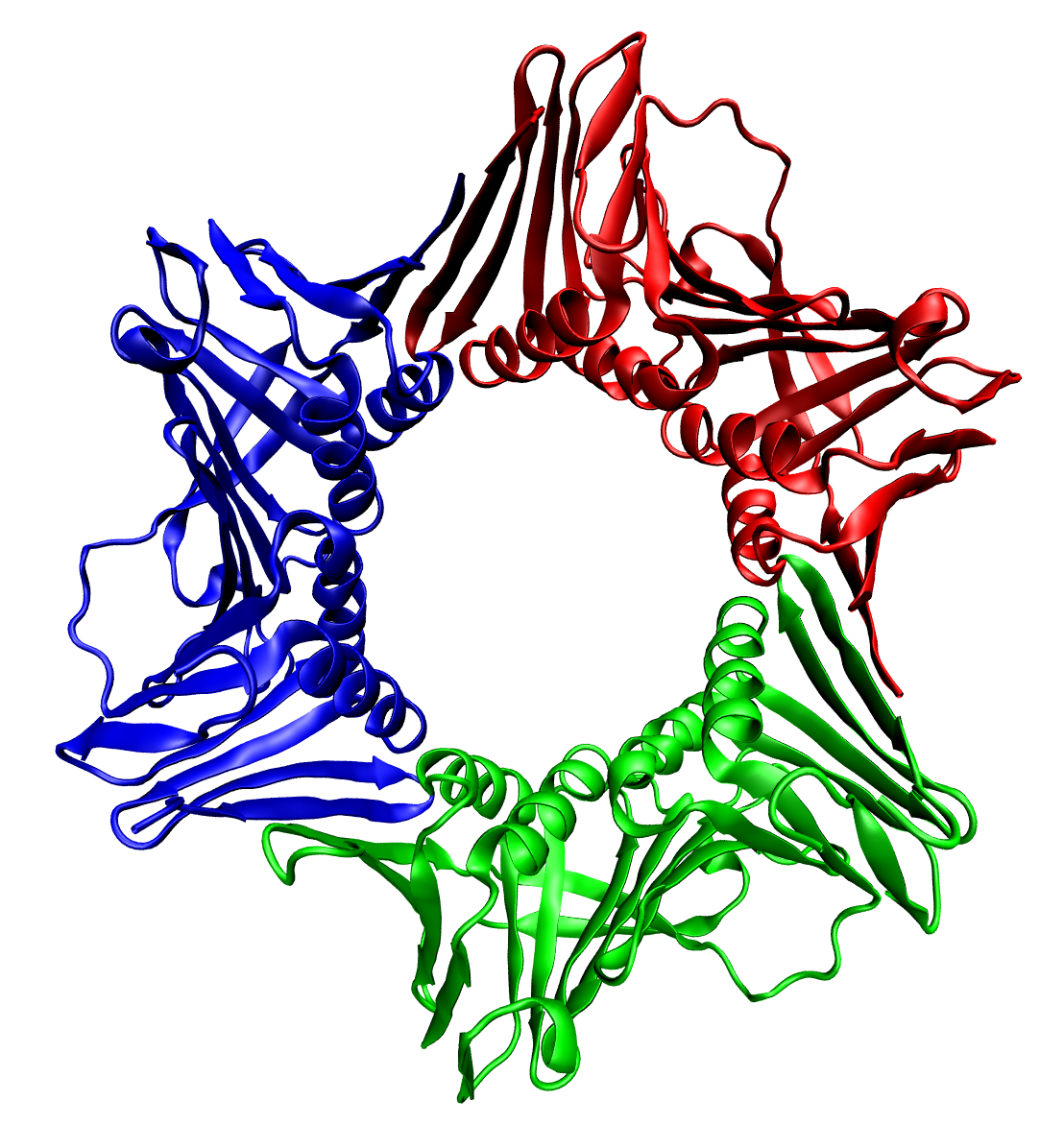
Protein Quaternary Structure
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors. Description and examples Many proteins are actually assemblies of multiple polypeptide chains. The quaternary structure refers to the number and arrangement of the protein subunits wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quaternary
The Quaternary ( ) is the current and most recent of the three period (geology), periods of the Cenozoic era (geology), Era in the geologic time scale of the International Commission on Stratigraphy (ICS). It follows the Neogene Period and spans from 2.58 million years ago to the present. The Quaternary Period is divided into two epochs: the Pleistocene (2.58 million years ago to 11.7 thousand years ago) and the Holocene (11.7 thousand years ago to today, although a third epoch, the Anthropocene, has been proposed but is not yet officially recognised by the ICS). The Quaternary Period is typically defined by the cyclic growth and decay of continental ice sheets related to the Milankovitch cycles and the associated climate and environmental changes that they caused. Research history In 1759 Giovanni Arduino (geologist), Giovanni Arduino proposed that the geological strata of northern Italy could be divided into four successive formations or "orders" ( it, quattro ord ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA, transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain vi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allostery
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. Allosteric regulation is also particularly important in the cell's ability to adjust enzyme activity. The term ''allostery'' comes from the Ancient Greek ''allos'' (), "other", and ''stere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cooperative Binding
Molecular binding is an interaction between molecules that results in a stable physical association between those molecules. Cooperative binding occurs in binding systems containing more than one type, or species, of molecule and in which one of the partners is not mono-valent and can bind more than one molecule of the other species. For example, consider a system where one molecule of species A can bind to molecules of species B. Species A is called the receptor and species B is called the ligand. Binding can be considered "cooperative" if the binding of the first molecule of B to A changes the binding affinity of the second B molecule, making it more or less likely to bind. In other words, the binding of B molecules to the different sites on A do not constitute mutually independent events. Cooperativity can be positive or negative. Cooperative binding is observed in many biopolymers, including proteins and nucleic acids. Cooperative binding has been shown to be the mechanism unde ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Conformation
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microtubule
Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement. Microtubules play an important role in a number of cellular processes. They are involved in maintaining the structure of the cell and, together with microfilaments and intermediate filaments, they form the cytoskeleton. They also make up the internal structure of cilia and flagella. They provide platforms for intracellular transport and are involved in a variety of cellular processes, including the movement of secretory vesicles, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Holoenzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction rat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |