|
Protein Aggregation
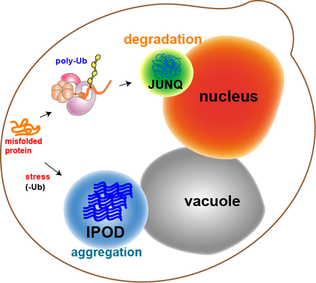
In molecular biology, protein aggregation is a phenomenon in which intrinsically-disordered or mis-folded proteins aggregate (i.e., accumulate and clump together) either intra- or extracellularly. Protein aggregates have been implicated in a wide variety of diseases known as amyloidoses, including ALS, Alzheimer's, Parkinson's and prion disease. After synthesis, proteins typically fold into a particular three-dimensional conformation that is the most thermodynamically favorable: their native state. This folding process is driven by the hydrophobic effect: a tendency for hydrophobic (water-fearing) portions of the protein to shield themselves from the hydrophilic (water-loving) environment of the cell by burying into the interior of the protein. Thus, the exterior of a protein is typically hydrophilic, whereas the interior is typically hydrophobic. Protein structures are stabilized by non-covalent interactions and disulfide bonds between two cysteine residues. The non-covale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Aggregation
In molecular biology, protein aggregation is a phenomenon in which intrinsically-disordered or mis-folded proteins aggregate (i.e., accumulate and clump together) either intra- or extracellularly. Protein aggregates have been implicated in a wide variety of diseases known as amyloidoses, including ALS, Alzheimer's, Parkinson's and prion disease. After synthesis, proteins typically fold into a particular three-dimensional conformation that is the most thermodynamically favorable: their native state. This folding process is driven by the hydrophobic effect: a tendency for hydrophobic (water-fearing) portions of the protein to shield themselves from the hydrophilic (water-loving) environment of the cell by burying into the interior of the protein. Thus, the exterior of a protein is typically hydrophilic, whereas the interior is typically hydrophobic. Protein structures are stabilized by non-covalent interactions and disulfide bonds between two cysteine residues. The non-covale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exceptions, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dipole-dipole Bond
An intermolecular force (IMF) (or secondary force) is the force that mediates interaction between molecules, including the electromagnetic forces of attraction or repulsion which act between atoms and other types of neighbouring particles, e.g. atoms or ions. Intermolecular forces are weak relative to intramolecular forces – the forces which hold a molecule together. For example, the covalent bond, involving sharing electron pairs between atoms, is much stronger than the forces present between neighboring molecules. Both sets of forces are essential parts of force fields frequently used in molecular mechanics. The investigation of intermolecular forces starts from macroscopic observations which indicate the existence and action of forces at a molecular level. These observations include non-ideal-gas thermodynamic behavior reflected by virial coefficients, vapor pressure, viscosity, superficial tension, and absorption data. The first reference to the nature of microscopic force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Such an interacting system is generally denoted , where the solid line denotes a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are the second-row elements nitrogen (N), oxygen (O), and fluorine (F). Hydrogen bonds can be intermolecular (occurring between separate molecules) or intramolecular (occurring among parts of the same molecule). The energy of a hydrogen bond depends on the geometry, the environment, and the nature of the specific donor and acceptor atoms and can vary between 1 and 40 kcal/mol. This makes them somewhat stronger than a van der Waals interaction, and weaker than fully covalent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
London Dispersion Force
London dispersion forces (LDF, also known as dispersion forces, London forces, instantaneous dipole–induced dipole forces, fluctuating induced dipole bonds or loosely as van der Waals forces) are a type of intermolecular force acting between atoms and molecules that are normally electrically symmetric; that is, the electrons are symmetrically distributed with respect to the nucleus. They are part of the van der Waals forces. The LDF is named after the German physicist Fritz London. They are the weakest intermolecular force. Introduction The electron distribution around an atom or molecule undergoes fluctuations in time. These fluctuations create instantaneous electric fields which are felt by other nearby atoms and molecules, which in turn adjust the spatial distribution of their own electrons. The net effect is that the fluctuations in electron positions in one atom induce a corresponding redistribution of electrons in other atoms, such that the electron motions become corre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Salt Bridge (protein And Supramolecular)
In chemistry, a salt bridge is a combination of two non-covalent interactions: hydrogen bonding and ionic bonding (Figure 1). Ion pairing is one of the most important noncovalent forces in chemistry, in biological systems, in different materials and in many applications such as ion pair chromatography. It is a most commonly observed contribution to the stability to the entropically unfavorable folded conformation of proteins. Although non-covalent interactions are known to be relatively weak interactions, small stabilizing interactions can add up to make an important contribution to the overall stability of a conformer. Not only are salt bridges found in proteins, but they can also be found in supramolecular chemistry. The thermodynamics of each are explored through experimental procedures to access the free energy contribution of the salt bridge to the overall free energy of the state. Salt bridges in chemical bonding In water, formation of salt bridges or ion pairs is most ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
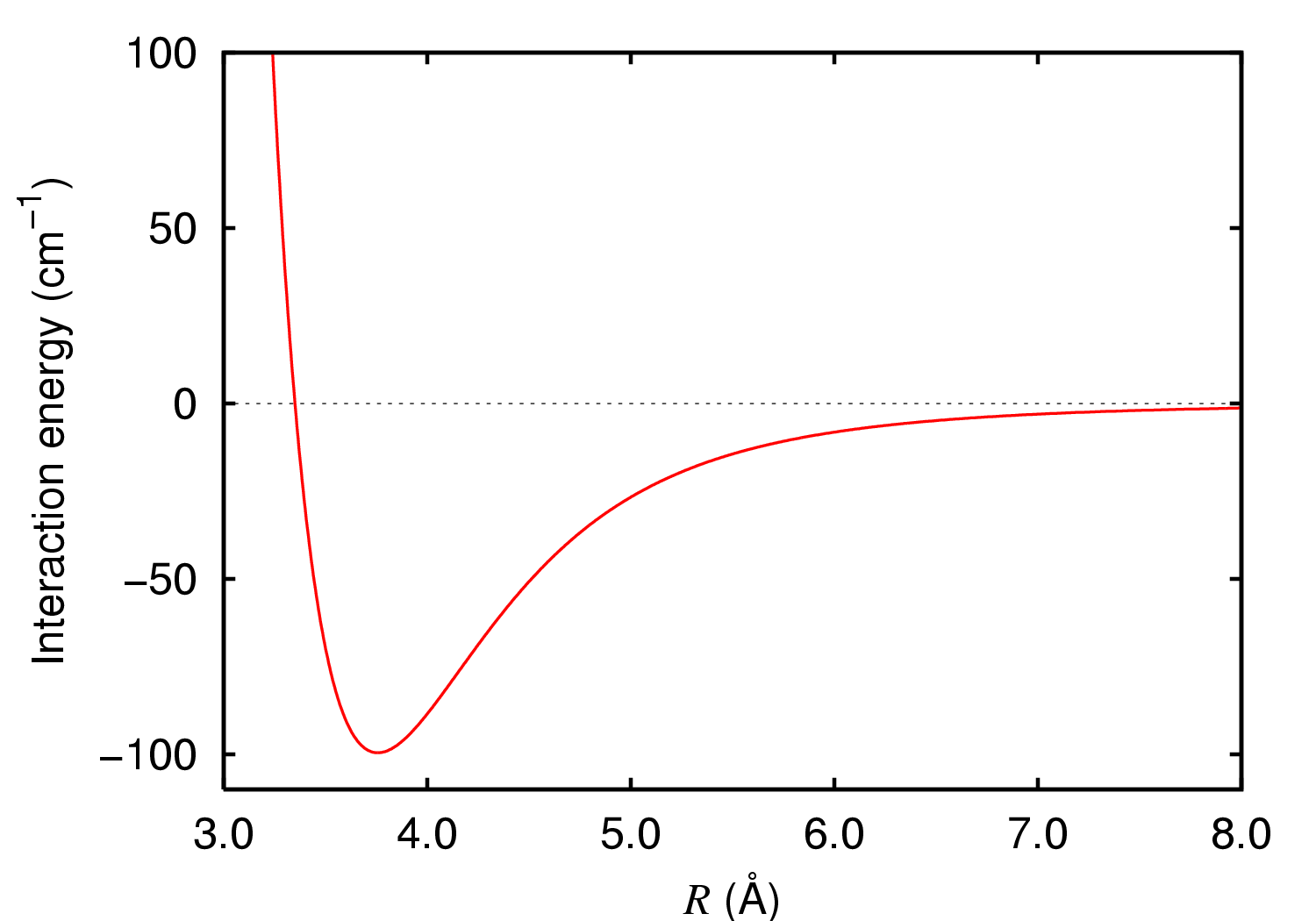
Van Der Waals Force
In molecular physics, the van der Waals force is a distance-dependent interaction between atoms or molecules. Unlike ionic or covalent bonds, these attractions do not result from a chemical electronic bond; they are comparatively weak and therefore more susceptible to disturbance. The van der Waals force quickly vanishes at longer distances between interacting molecules. Named after Dutch physicist Johannes Diderik van der Waals, the van der Waals force plays a fundamental role in fields as diverse as supramolecular chemistry, structural biology, polymer science, nanotechnology, surface science, and condensed matter physics. It also underlies many properties of organic compounds and molecular solids, including their solubility in polar and non-polar media. If no other force is present, the distance between atoms at which the force becomes repulsive rather than attractive as the atoms approach one another is called the van der Waals contact distance; this phenomenon resul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ionic Bonding
Ionic bonding is a type of chemical bonding that involves the electrostatic attraction between oppositely charged ions, or between two atoms with sharply different electronegativities, and is the primary interaction occurring in ionic compounds. It is one of the main types of bonding along with covalent bonding and metallic bonding. Ions are atoms (or groups of atoms) with an electrostatic charge. Atoms that gain electrons make negatively charged ions (called anions). Atoms that lose electrons make positively charged ions (called cations). This transfer of electrons is known as electrovalence in contrast to covalence. In the simplest case, the cation is a metal atom and the anion is a nonmetal atom, but these ions can be of a more complex nature, e.g. molecular ions like or . In simpler words, an ionic bond results from the transfer of electrons from a metal to a non-metal in order to obtain a full valence shell for both atoms. It is important to recognize that ''clean'' ion ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |





_mutation_R527L_PMID_22549407_surface_and_cartoon.png)


