|
Prp24
Prp24 (precursor RNA processing, gene 24) is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to ''E. coli'', fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing. Biological Role The process of spliceosome formation involves the U4 and U6 snRNPs associating and forming a di-snRNP in the cell nucleus. This di-snRNP then recruits another member ( U5) to become a tri-snRNP. U6 must then dissociate from U4 to bond with U2 and become catalytically active. Once splicing has been done, U6 must dissociate from the spliceosome and bond back with U4 to restart the cycle. Prp24 has been shown ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
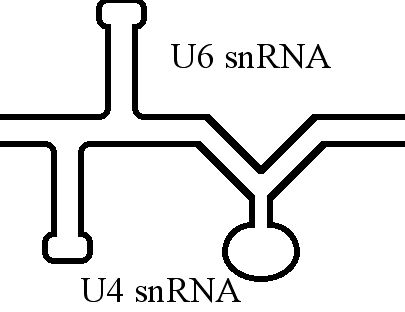
U6 SnRNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many organism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U4 SnRNA
The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved. Biological role The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNP, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been proposed to coincide with different temp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnRNP
snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. Additionally, '' U7 snRNP'' is not involved in splicing at all, as U7 snRNP is responsible for processing the 3′ stem-loop of histone pre-mRNA. The two essential components of snRNPs are protein molecules and RNA. The RNA found within each snRNP particle is known as ''small nuclear RNA'', or snRNA, and is usually about 150 nucleotides in length. The snRNA component of the snRNP gives specificity to individual introns by " recognizing" the sequences of critical splicing signals at the 5' and 3' ends and branch site of introns. The snRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HnRNA
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing. Pre-mRNA is synthesized from a DNA template in the cell nucleus by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely processed, it is termed "mature messenger RNA", or simply "messenger RNA". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA. There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and compl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Sequence
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, prompt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dissociation Constant
In chemistry, biochemistry, and pharmacology, a dissociation constant (K_D) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into its component ions. The dissociation constant is the inverse of the association constant. In the special case of salts, the dissociation constant can also be called an ionization constant. For a general reaction: : A_\mathit B_\mathit \mathit A + \mathit B in which a complex \ce_x \ce_y breaks down into ''x'' A subunits and ''y'' B subunits, the dissociation constant is defined as : K_D = \frac where and ''x'' B''y''are the equilibrium concentrations of A, B, and the complex A''x'' B''y'', respectively. One reason for the popularity of the dissociation constant in biochemistry and pharmacology is that in the frequently encount ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta-sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substrate (biochemistry)
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed. In the former sense, a reagent is added to the ''substrate'' to generate a product through a chemical reaction. The term is used in a similar sense in synthetic and organic chemistry, where the substrate is the chemical of interest that is being modified. In biochemistry, an enzyme substrate is the material upon which an enzyme acts. When referring to Le Chatelier's principle, the substrate is the reagent whose concentration is changed. ;Spontaneous reaction : :*Where S is substrate and P is product. ;Catalysed reaction : :*Where S is substrate, P is product and C is catalyst. In the latter sense, it may refer to a surface on which other chemical reactions are performed or play a supporting role in a variety of spectroscopic and microscop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Annealing (biology)
Nucleic acid thermodynamics is the study of how temperature affects the nucleic acid structure of double-stranded DNA (dsDNA). The melting temperature (''Tm'') is defined as the temperature at which half of the DNA strands are in the random coil or single-stranded (ssDNA) state. ''Tm'' depends on the length of the DNA molecule and its specific nucleotide sequence. DNA, when in a state where its two strands are dissociated (i.e., the dsDNA molecule exists as two independent strands), is referred to as having been denatured by the high temperature. Concepts Hybridization Hybridization is the process of establishing a non-covalent, sequence-specific interaction between two or more complementary strands of nucleic acids into a single complex, which in the case of two strands is referred to as a duplex. Oligonucleotides, DNA, or RNA will bind to their complement under normal conditions, so two perfectly complementary strands will bind to each other readily. In order to reduce the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Weight
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic bonds, are typically not consi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |