|
Primer Extension
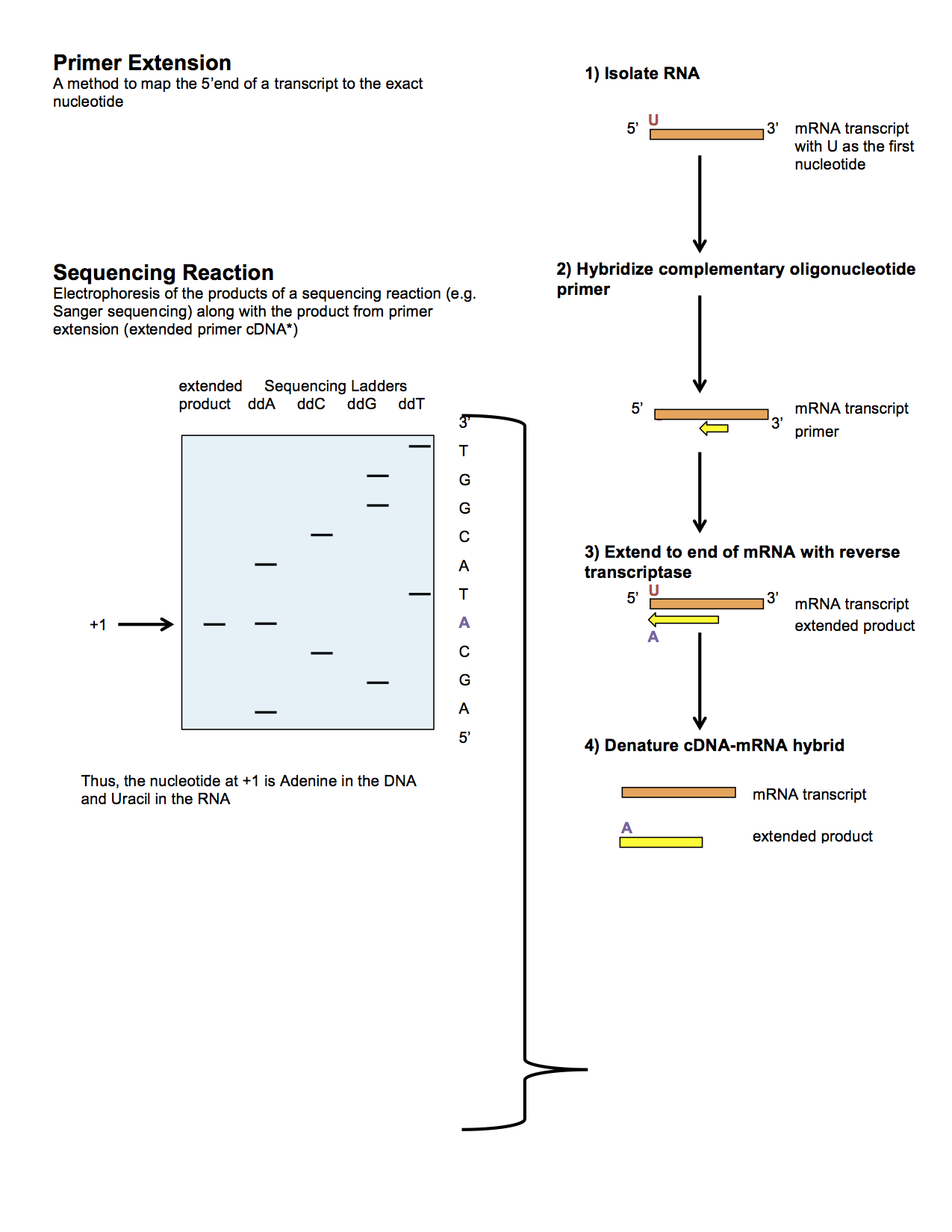
Primer extension is a technique whereby the 5' ends of RNA can be mapped - that is, they can be sequenced and properly identified. Primer extension can be used to determine the start site of transcription (the end site cannot be determined by this method) by which its sequence is known. This technique requires a radiolabelled primer (usually 20 - 50 nucleotides in length) which is complementary to a region near the 3' end of the mRNA. The primer is allowed to anneal to the RNA and reverse transcriptase is used to synthesize cDNA from the RNA until it reaches the 5' end of the RNA. By denaturing the hybrid and using the extended primer cDNA as a marker on an electrophoretic gel, it is possible to determine the transcriptional start site. It is usually done so by comparing its location on the gel with the DNA sequence (e.g. Sanger sequencing), preferably by using the same primer on the DNA template strand. The exact nucleotide by which the transcription starts at can be pinpointe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primer Extension Assay
Primer may refer to: Arts, entertainment, and media Films * ''Primer'' (film), a 2004 feature film written and directed by Shane Carruth * ''Primer'' (video), a documentary about the funk band Living Colour Literature * Primer (textbook), a textbook used in primary education to teach the alphabet and other basic subjects * Primer (prayer book), a common name for English prayer books used from the 13th to 16th centuries * ''The New England Primer'' (1688), a Puritan book from Colonial America with morality-themed rhymes Music * ''Primer'' (album), a 1995 music album by the musical group Rockapella * Primer 55, an American alternative metal band * "The Primer", a song from the 2005 album ''Alaska'' by Between the Buried and Me Firearms * Primer (firearms), a firearm powder charge-ignition mechanism ** Centerfire ammunition, Boxer or Berdan primers used in modern centerfire cartridges ** Detonator, a small explosive device also known as an explosive primer or blasting cap ** Fri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5' End
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to an mR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Start Site
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sanger Sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. It was first commercialized by Applied Biosystems in 1986. More recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies (like Illumina) in that it can produce DNA sequence reads of > 500 nucleotides and maintains a very low error rate with accuracies around 99.99%. Sanger sequencing is still actively being used in efforts for public health initiative ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclease Protection Assay
Nuclease protection assay is a laboratory technique used in biochemistry and genetics to identify individual RNA molecules in a heterogeneous RNA sample extracted from cells. The technique can identify one or more RNA molecules of known sequence even at low total concentration. The extracted RNA is first mixed with antisense RNA or DNA probes that are complementary to the sequence or sequences of interest and the complementary strands are hybridized to form double-stranded RNA (or a DNA-RNA hybrid). The mixture is then exposed to ribonucleases that specifically cleave only ''single''-stranded RNA but have no activity against double-stranded RNA. When the reaction runs to completion, susceptible RNA regions are degraded to very short oligomers or to individual nucleotides; the surviving RNA fragments are those that were complementary to the added antisense strand and thus contained the sequence of interest. Probe The probes are prepared by cloning part of the gene of intere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hybridization Probe
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA of usually 15–10000 nucleotide long which can be radioactively or fluorescently labeled. HP can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA ( Southern blotting) or RNA (northern blotting) immobilized on a membrane or in situ. To detect hybridization of the probe to its target sequence, the probe is tagged (or "labeled") with a molecular marker of either radioactive or (more recently) fluorescent molecules. Commonly used markers are 32P (a radioactive isotope of phosphorus incorporated into the phosphodiester bond in the probe DNA), digoxigenin, a non-radioactive, antibody-based marker, biotin or fluorescein. DNA sequences o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S1 Mapping
Nuclease protection assay is a laboratory technique used in biochemistry and genetics to identify individual RNA molecules in a heterogeneous RNA sample extracted from cells. The technique can identify one or more RNA molecules of known sequence even at low total concentration. The extracted RNA is first mixed with antisense RNA or DNA probes that are complementary to the sequence or sequences of interest and the complementary strands are hybridized to form double-stranded RNA (or a DNA-RNA hybrid). The mixture is then exposed to ribonucleases that specifically cleave only ''single''-stranded RNA but have no activity against double-stranded RNA. When the reaction runs to completion, susceptible RNA regions are degraded to very short oligomers or to individual nucleotides; the surviving RNA fragments are those that were complementary to the added antisense strand and thus contained the sequence of interest. Probe The probes are prepared by cloning part of the gene of interest ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S1 Nuclease
Nuclease S1 () is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalysis, catalyses the following chemical reaction : Endonucleolytic cleavage to 5'-phosphomononucleotide and 5'-phosphooligonucleotide end-products Although its primary substrate is single-stranded, it can also occasionally introduce single-stranded breaks in double-stranded DNA or RNA, or DNA-RNA hybrids. The enzyme hydrolyses single stranded region in duplex DNA such as loops or gaps. It also cleaves a strand opposite a nick on the complementary strand. It has no sequence specificity. Well-known versions include S1 found in ''Aspergillus oryzae'' (yellow koji mold) and Nuclease P1 found in ''Penicillium citrinum''. Members of the S1/P1 family are found in both prokaryotes and eukaryotes and are thought to be associated in programmed cell death and also in tissue differentiation. Furthermore, they are secretion, secreted extracellular, that is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |