|
PREDITOR
PREDITOR (PREDIction of TORsion angles) is a freely available web-server for the prediction of protein torsion angles from chemical shifts. For many years it has been known that protein chemical shifts are sensitive to protein secondary structure, which in turn, is sensitive to backbone torsion angles. torsion angles are internal coordinates that can be used to describe the conformation of a polypeptide chain. They can also be used as constraints to help determine or refine protein structures via NMR spectroscopy. In proteins there are four major torsion angles of interest: phi, psi, omega and chi-1. Traditionally protein NMR spectroscopists have used vicinal J-coupling information and the Karplus relation to determine approximate backbone torsion angle constraints for phi and chi-1 angles. However, several studies in the early 1990s pointed out the strong relationship between 1H and 13C chemical shifts and torsion angles, especially with backbone phi and psi angles. Later a num ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
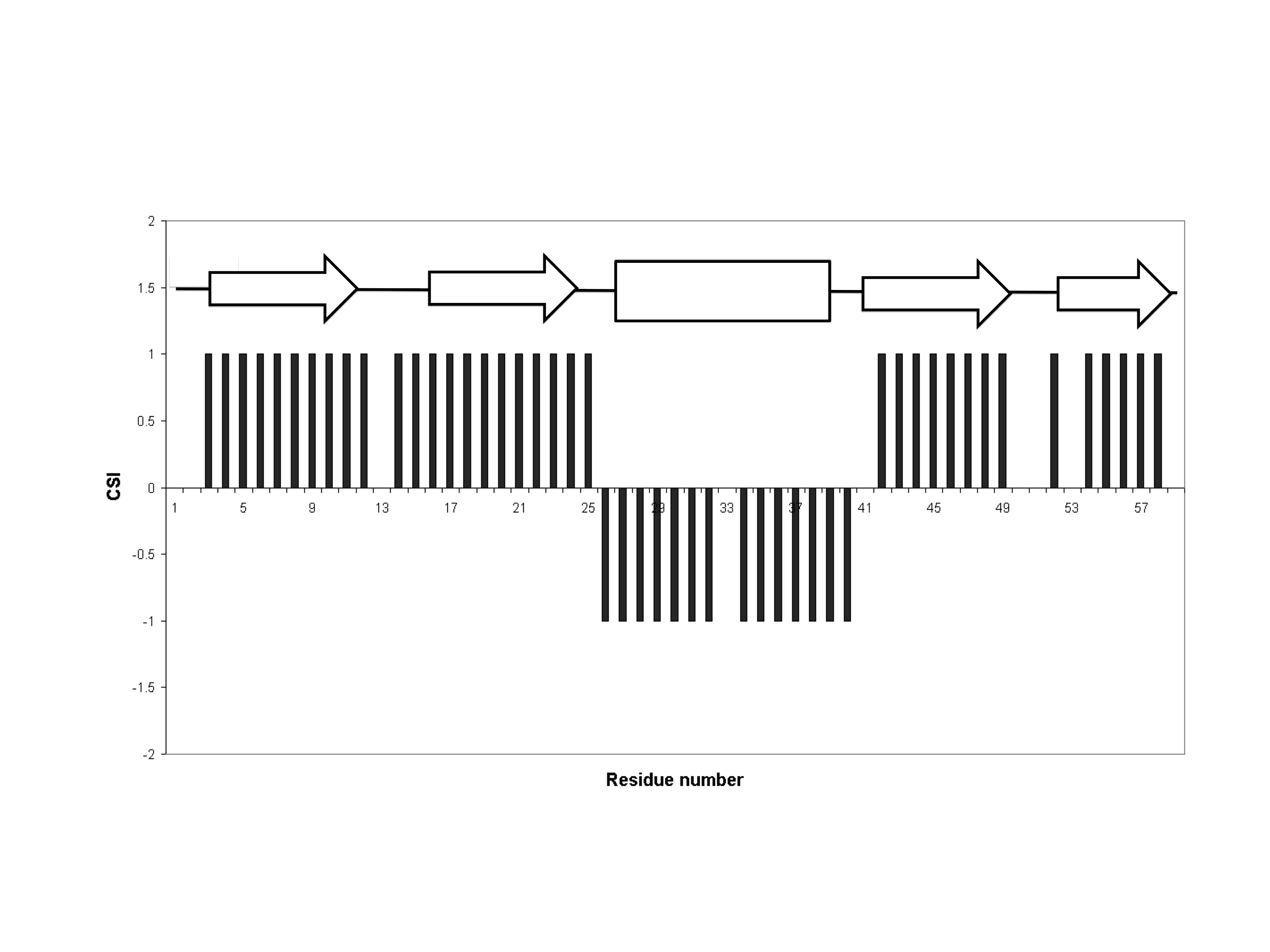
Chemical Shift Index
The chemical shift index or CSI is a widely employed technique in protein nuclear magnetic resonance spectroscopy that can be used to display and identify the location (i.e. start and end) as well as the type of protein secondary structure (beta strands, helices and random coil regions) found in proteins using only backbone chemical shift data The technique was invented by David S. Wishart in 1992 for analyzing 1Hα chemical shifts and then later extended by him in 1994 to incorporate 13C backbone shifts. The original CSI method makes use of the fact that 1Hα chemical shifts of amino acid residues in helices tends to be shifted upfield (i.e. towards the right side of an NMR spectrum) relative to their random coil values and downfield (i.e. towards the left side of an NMR spectrum) in beta strands. Similar kinds of upfield and downfield trends are also detectable in backbone 13C chemical shifts. Implementation The CSI is a graph-based technique that essentially employs an amino ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
David S
David (; , "beloved one") (traditional spelling), , ''Dāwūd''; grc-koi, Δαυΐδ, Dauíd; la, Davidus, David; gez , ዳዊት, ''Dawit''; xcl, Դաւիթ, ''Dawitʿ''; cu, Давíдъ, ''Davidŭ''; possibly meaning "beloved one". was, according to the Hebrew Bible, the third king of the United Kingdom of Israel. In the Books of Samuel, he is described as a young shepherd and harpist who gains fame by slaying Goliath, a champion of the Philistines, in southern Canaan. David becomes a favourite of Saul, the first king of Israel; he also forges a notably close friendship with Jonathan, a son of Saul. However, under the paranoia that David is seeking to usurp the throne, Saul attempts to kill David, forcing the latter to go into hiding and effectively operate as a fugitive for several years. After Saul and Jonathan are both killed in battle against the Philistines, a 30-year-old David is anointed king over all of Israel and Judah. Following his rise to power, David ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Torsion Angle
A dihedral angle is the angle between two intersecting planes or half-planes. In chemistry, it is the clockwise angle between half-planes through two sets of three atoms, having two atoms in common. In solid geometry, it is defined as the union of a line and two half-planes that have this line as a common edge. In higher dimensions, a dihedral angle represents the angle between two hyperplanes. The planes of a flying machine are said to be at positive dihedral angle when both starboard and port main planes (commonly called wings) are upwardly inclined to the lateral axis. When downwardly inclined they are said to be at a negative dihedral angle. Mathematical background When the two intersecting planes are described in terms of Cartesian coordinates by the two equations : a_1 x + b_1 y + c_1 z + d_1 = 0 :a_2 x + b_2 y + c_2 z + d_2 = 0 the dihedral angle, \varphi between them is given by: :\cos \varphi = \frac and satisfies 0\le \varphi \le \pi/2. Alternatively, if and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Shift
In nuclear magnetic resonance (NMR) spectroscopy, the chemical shift is the resonant frequency of an atomic nucleus relative to a standard in a magnetic field. Often the position and number of chemical shifts are diagnostic of the structure of a molecule. Chemical shifts are also used to describe signals in other forms of spectroscopy such as photoemission spectroscopy. Some atomic nuclei possess a magnetic moment ( nuclear spin), which gives rise to different energy levels and resonance frequencies in a magnetic field. The total magnetic field experienced by a nucleus includes local magnetic fields induced by currents of electrons in the molecular orbitals (note that electrons have a magnetic moment themselves). The electron distribution of the same type of nucleus (e.g. ) usually varies according to the local geometry (binding partners, bond lengths, angles between bonds, and so on), and with it the local magnetic field at each nucleus. This is reflected in the spin energy le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NMR Spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique to observe local magnetic fields around atomic nuclei. The sample is placed in a magnetic field and the NMR signal is produced by excitation of the nuclei sample with radio waves into nuclear magnetic resonance, which is detected with sensitive radio receivers. The intramolecular magnetic field around an atom in a molecule changes the resonance frequency, thus giving access to details of the electronic structure of a molecule and its individual functional groups. As the fields are unique or highly characteristic to individual compounds, in modern organic chemistry practice, NMR spectroscopy is the definitive method to identify monomolecular organic compounds. The principle of NMR usually involves three sequential steps: # The alignment (polarization) of the magnetic nuclear spins in an applied, constant magnetic field B0. # The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Shift
In nuclear magnetic resonance (NMR) spectroscopy, the chemical shift is the resonant frequency of an atomic nucleus relative to a standard in a magnetic field. Often the position and number of chemical shifts are diagnostic of the structure of a molecule. Chemical shifts are also used to describe signals in other forms of spectroscopy such as photoemission spectroscopy. Some atomic nuclei possess a magnetic moment ( nuclear spin), which gives rise to different energy levels and resonance frequencies in a magnetic field. The total magnetic field experienced by a nucleus includes local magnetic fields induced by currents of electrons in the molecular orbitals (note that electrons have a magnetic moment themselves). The electron distribution of the same type of nucleus (e.g. ) usually varies according to the local geometry (binding partners, bond lengths, angles between bonds, and so on), and with it the local magnetic field at each nucleus. This is reflected in the spin energy le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary structures ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein NMR
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how rapid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |