|
PAP1
Retinitis pigmentosa 9 (autosomal dominant), also known as RP9 or PAP-1, is a protein which in humans is encoded by the ''RP9'' gene. Function The removal of introns from nuclear pre-mRNAs occurs on a complex called a spliceosome, which is made up of 4 small nuclear ribonucleoprotein (snRNP) particles and an undefined number of transiently associated splicing factors. The exact role of PAP-1 in splicing is not fully understood, but it is thought that PAP-1 localizes in nuclear speckles containing the splicing factor SC35 and interacts directly with another splicing factor, U2AF35. Clinical significance Mutations in PAP1 underlie autosomal dominant retinitis pigmentosa mapped to the RP9 gene locus. Interactions RP9 has been shown to interact with U2 small nuclear RNA auxiliary factor 1 Splicing factor U2AF 35 kDa subunit is a protein that in humans is encoded by the ''U2AF1'' gene. Function This gene belongs to the splicing factor SR family of genes . U2AF1 is a subu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Retinitis Pigmentosa
Retinitis pigmentosa (RP) is a genetic disorder of the eyes that causes loss of vision. Symptoms include trouble seeing at night and decreasing peripheral vision (side and upper or lower visual field). As peripheral vision worsens, people may experience "tunnel vision". Complete blindness is uncommon. Onset of symptoms is generally gradual and often begins in childhood. Retinitis pigmentosa is generally inherited from one or both parents or rarely it can be caused by a miscoding during DNA division. It is caused by genetic miscoding of proteins in one of more than 300 genes involved. The underlying mechanism involves the progressive loss of rod photoreceptor cells that line the retina of the eyeball. The rod cells secrete a neuroprotective substance (Rod-derived cone viability factor, RdCVF) that protects the cone cells from apoptosis (cell death). However, when the rod cells die, this substance is no longer provided. This is generally followed by the loss of cone photorecept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRNA introns, group I introns, group II introns, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced “snurps”), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut. However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes. History In 1977, work by the Sharp and Roberts labs reveale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnRNP
snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. Additionally, '' U7 snRNP'' is not involved in splicing at all, as U7 snRNP is responsible for processing the 3′ stem-loop of histone pre-mRNA. The two essential components of snRNPs are protein molecules and RNA. The RNA found within each snRNP particle is known as ''small nuclear RNA'', or snRNA, and is usually about 150 nucleotides in length. The snRNA component of the snRNP gives specificity to individual introns by " recognizing" the sequences of critical splicing signals at the 5' and 3' ends and branch site of introns. The snRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SFRS2
Splicing factor, arginine/serine-rich 2 is a protein that in humans is encoded by the ''SFRS2'' gene. MDS-associated splicing factor SRSF2 affects the expression of Class III and Class IV isoforms and perturbs granulopoiesis and SRSF2 P95H promotes Class IV splicing by binding to key ESE sequences in CSF3R exon 17, and that SRSF2, when mutated, contributes to dysgranulopoiesis. Interactions SFRS2 has been shown to interact with CDC5L and ASF/SF2 Serine/arginine-rich splicing factor 1 (SRSF1) also known as alternative splicing factor 1 (ASF1), pre-mRNA-splicing factor SF2 (SF2) or ASF1/SF2 is a protein that in humans is encoded by the ''SRSF1'' gene. ASF/SF2 is an essential sequence specif .... References Further reading * * * * * * * * * * * * * * * * * * External links * {{gene-17-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U2 Small Nuclear RNA Auxiliary Factor 1
Splicing factor U2AF 35 kDa subunit is a protein that in humans is encoded by the ''U2AF1'' gene. Function This gene belongs to the splicing factor SR family of genes . U2AF1 is a subunit of the U2 Auxiliary Factor complex alongside a larger subunit, U2AF2. U2AF1 is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. This gene encodes a small (~35 kDa) subunit which plays a critical role in RNA splicing by recognizing and binding to AG nucleotides at the 3’ splice site to facilitate spliceosome assembly. Alternatively spliced transcript variants encoding different isoforms have been identified . Somatic mutations in ''U2AF1'' have been found in a range of human cancers, with a distinctive pattern of these mutations at the zinc fingers implicating a functional role under selection. In lung cancers, these mutations affect alternative splicing of several transcripts, including oncogenic ROS1 fusions. U2af1 conditional deletion in mouse hematopoi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autosomal Dominant
In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (''de novo'') or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes ( autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child (see Sex linkage). Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance such as incomplete d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
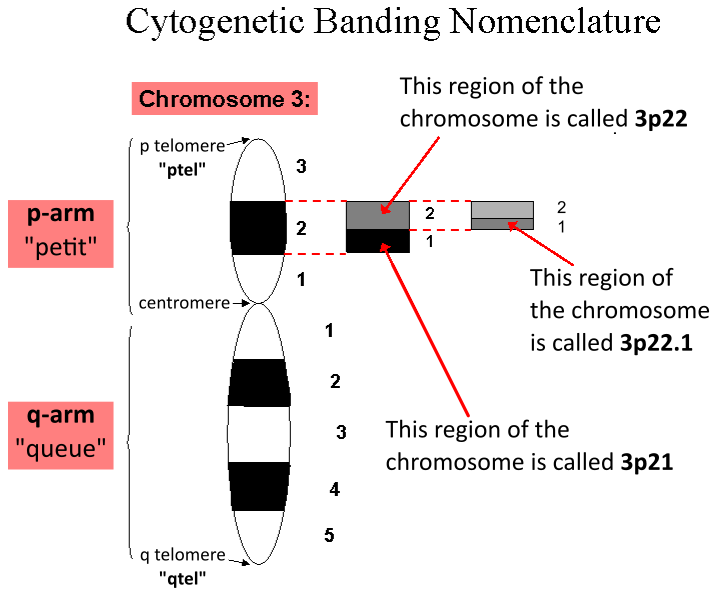
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |