|
PAK3
PAK3 (p21-activated kinase 2, beta-PAK) is one of three members of Group I PAK family of evolutionary conserved serine/threonine kinases. PAK3 is preferentially expressed in neuronal cells and involved in synapse formation and plasticity and mental retardation. Discovery PAK3 was initially cloned from a murine fibroblast cDNA library and from a murine embryo cDNA library. Like other group I PAKs, PAK3 is stimulated by activated Cdc42 and Rac1. Gene and spliced variants The human PAK3 gene, the longest group I family member, is 283-kb long. The PAK3 gene is composed of 22 exons of which 6 exons are for 5’-UTR and generates 13 alternative spliced transcripts. Among PAK3 transcripts, 11 transcripts are for coding proteins ranging from 181- to 580-amino acids long, while remaining two transcripts are non-coding RNAs. The murine PAK3 gene contains 10 transcripts, coding six proteins from 544 amino acids and 559 amino acids long, and four smaller polypeptides from 23 to 366 a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAK1
Serine/threonine-protein kinase PAK 1 is an enzyme that in humans is encoded by the ''PAK1'' gene. PAK1 is one of six members of the PAK family of serine/threonine kinases which are broadly divided into group I (PAK1, PAK2 and PAK3) and group II (PAK4, PAK6 and PAK5/7). The PAKs are evolutionarily conserved. PAK1 localizes in distinct sub-cellular domains in the cytoplasm and nucleus. PAK1 regulates cytoskeleton remodeling, phenotypic signaling and gene expression, and affects a wide variety of cellular processes such as directional motility, invasion, metastasis, growth, cell cycle progression, angiogenesis. PAK1-signaling dependent cellular functions regulate both physiologic and disease processes, including cancer, as PAK1 is widely overexpressed and hyperstimulated in human cancer, at-large. Discovery PAK1 was first discovered as an effector of the Rho GTPases in rat brain by Manser and colleagues in 1994. The human PAK1 was identified as a GTP-dependent interacting partner ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine/threonine Kinases
A serine/threonine protein kinase () is a kinase enzyme, in particular a protein kinase, that phosphorylates the OH group of the amino-acid residues serine or threonine, which have similar side chains. At least 350 of the 500+ human protein kinases are serine/threonine kinases (STK). In enzymology, the term ''serine/threonine protein kinase'' describes a class of enzymes in the family of transferases, that transfer phosphates to the oxygen atom of a serine or threonine side chain in proteins. This process is called phosphorylation. Protein phosphorylation in particular plays a significant role in a wide range of cellular processes and is a very important posttranslational modification. The chemical reaction performed by these enzymes can be written as :ATP + a protein \rightleftharpoons ADP + a phosphoprotein Thus, the two substrates of this enzyme are ATP and a protein, whereas its two products are ADP and phosphoprotein. The systematic name of this enzyme class is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synapse
In the nervous system, a synapse is a structure that permits a neuron (or nerve cell) to pass an electrical or chemical signal to another neuron or to the target effector cell. Synapses are essential to the transmission of nervous impulses from one neuron to another. Neurons are specialized to pass signals to individual target cells, and synapses are the means by which they do so. At a synapse, the plasma membrane of the signal-passing neuron (the ''presynaptic'' neuron) comes into close apposition with the membrane of the target (''postsynaptic'') cell. Both the presynaptic and postsynaptic sites contain extensive arrays of molecular machinery that link the two membranes together and carry out the signaling process. In many synapses, the presynaptic part is located on an axon and the postsynaptic part is located on a dendrite or soma. Astrocytes also exchange information with the synaptic neurons, responding to synaptic activity and, in turn, regulating neurotransmission. Syna ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intellectual Disability
Intellectual disability (ID), also known as general learning disability in the United Kingdom and formerly mental retardation,Rosa's Law, Pub. L. 111-256124 Stat. 2643(2010). is a generalized neurodevelopmental disorder characterized by significantly impaired intellectual and adaptive functioning. It is defined by an IQ under 70, in addition to deficits in two or more adaptive behaviors that affect everyday, general living. Intellectual functions are defined under DSM-V as reasoning, problem‑solving, planning, abstract thinking, judgment, academic learning, and learning from instruction and experience, and practical understanding confirmed by both clinical assessment and standardized tests. Adaptive behavior is defined in terms of conceptual, social, and practical skills involving tasks performed by people in their everyday lives. Intellectual disability is subdivided into syndromic intellectual disability, in which intellectual deficits associated with other medical and be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
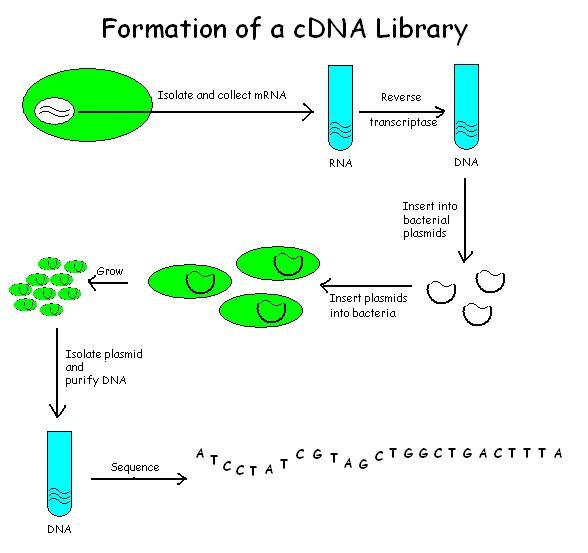
CDNA Library
A cDNA library is a combination of cloned cDNA (complementary DNA) fragments inserted into a collection of host cells, which constitute some portion of the transcriptome of the organism and are stored as a "library". cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue-specific cDNA libraries can be produced. In eukaryotic cells the mature mRNA is already spliced, hence the cDNA produced lacks introns and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library. cDNA Library Construction cDNA is created from a mature mRNA from a eukaryotic cell with the use of reverse transcriptase. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent transcriptomic and bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes referred to as "junk RNA"), spurious transcriptions. Others, however, disagree, arguing instead that many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AP-1 Transcription Factor
Activator protein 1 (AP-1) is a transcription factor that regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections. AP-1 controls a number of cellular processes including differentiation, proliferation, and apoptosis. The structure of AP-1 is a heterodimer composed of proteins belonging to the c-Fos, c-Jun, ATF and JDP families. History AP-1 was first discovered as a TPA-activated transcription factor that bound to a cis-regulatory element of the human metallothionein IIa ( hMTIIa) promoter and SV40. The AP-1 binding site was identified as the 12-O-Tetradecanoylphorbol-13-acetate ( TPA) response element (TRE) with the consensus sequence 5’-TGA G/C TCA-3’. The AP-1 subunit Jun was identified as a novel oncoprotein of avian sarcoma virus, and Fos-associated p39 protein was identified as the transcript of the cellular Jun gene. Fos was first isolated as the cellular homologue of two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |