|
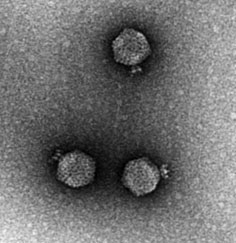
P1 Phage
P1 is a temperate bacteriophage that infects ''Escherichia coli'' and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages (e.g. the lambda phage) that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Temperateness (virology)
In virology, temperate refers to the ability of some bacteriophages (notably coliphage λ) to display a lysogenic life cycle. Many (but not all) temperate phages can integrate their genomes into their host bacterium's chromosome, together becoming a lysogen as the phage genome becomes a prophage. A temperate phage is also able to undergo a productive, typically lytic life cycle, where the prophage is expressed, replicates the phage genome, and produces phage progeny, which then leave the bacterium. With phage the term virulent is often used as an antonym to temperate, but more strictly a virulent phage is one that has lost its ability to display lysogeny through mutation rather than a phage lineage with no genetic potential to ever display lysogeny (which more properly would be described as an obligately lytic phage). Induction At some point, temperate bacteriophages switch from the lysogenic life cycle to the lytic life cycle. This conversion may happen spontaneously, althoug ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ff Phages
Ff phages (for ''F'' specific ''f''ilamentous phages) is a group of almost identical filamentous phage (genus ''Inovirus'') including phages f1, fd, M13 and ZJ/2, which infect bacteria bearing the F fertility factor. The virion (virus particle) is a flexible filament measuring about 6 by 900 nm, comprising a cylindrical protein tube protecting a single-stranded circular DNA molecule at its core. The phage codes for only 11 gene products, and is one of the simplest viruses known. It has been widely used to study fundamental aspects of molecular biology. George Smith and Greg Winter used f1 and fd for their work on phage display for which they were awarded a share of the 2018 Nobel Prize in Chemistry. Early experiments on Ff phages used M13 to identify gene functions, and M13 was also developed as a cloning vehicle, so the name M13 is sometimes used as an informal synonym for the whole group of Ff phages. Structure The virion is a flexible filament (worm-like chain) about 6&nb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-specific Recombination
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs. Site-specific recombination systems are highly specific, fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nat Sternberg
Nat L. Sternberg (August 2, 1942 – September 26, 1995) was an American molecular biology, molecular biologist and bacteriophage researcher, particularly known for his work on Genetic recombination, DNA recombination and the P1 phage, phage P1. Early life and education Born in 1942 in Brooklyn, New York (state), New York, Sternberg gained a BSc at Brooklyn College, followed by a master's at Long Island University. In 1969, he received a PhD from Purdue University, Indiana, researching T-even bacteriophages, T-even phage head proteins under the supervision of Sewell Champe. Career In 1970–72, Sternberg held a postdoctoral fellowship at the Laboratory of Molecular Biology of the Centre national de la recherche scientifique (CNRS) in Paris, under the direction of François Gros, where he researched the head proteins of Lambda phage, phage λ. He then returned to the US, taking up the position of staff fellow in the laboratory of Robert Weisberg at the National Institutes of Health ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Annual Review Of Virology
The ''Annual Review of Virology'' is an annual peer-reviewed scientific journal published by Annual Reviews. It was established in 2014 by editor Lynn W. Enquist (Princeton University). The journal covers all aspects of virology. History The ''Annual Review of Virology'' was first published in 2014 by nonprofit publisher Annual Reviews. By this time, Annual Reviews published over forty journal titles covering different disciplines; while more than half of the existing journal titles contained review articles relevant to virology, developments in the field were not routinely covered. Published articles on the study of viruses were not easy for readers to locate, as they were decentralized in the other discipline-specific journals. Lynn W. Enquist was the founding editor. Though it was initially published in print, as of 2021 it is only published electronically. Scope and indexing It defines its scope as covering significant developments in the study of all viruses, including t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Salvador Luria
Salvador Edward Luria (August 13, 1912 – February 6, 1991) was an Italian microbiologist, later a naturalized U.S. citizen. He won the Nobel Prize in Physiology or Medicine in 1969, with Max Delbrück and Alfred Hershey, for their discoveries on the replication mechanism and the genetic structure of viruses. Salvador Luria also showed that bacterial resistance to viruses (phages) is genetically inherited. Biography Early life Luria was born Salvatore Edoardo Luria in Turin, Italy to an influential Italian Sephardi Jewish family. His parents were Davide and Ester (Sacerdote) Luria. He attended the medical school at the University of Turin studying with Giuseppe Levi. There, he met two other future Nobel laureates: Rita Levi-Montalcini and Renato Dulbecco. He graduated from the University of Turin in 1935 and never got a master's degree or a PhD as they were not contemplated by the Italian high educational system (which, on the other hand, was very selective). From 1936 to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mu Phage
Bacteriophage Mu, also known as mu phage or mu bacteriophage, is a muvirus (the first of its kind to be identified) of the family ''Myoviridae'' which has been shown to cause genetic transposition. It is of particular importance as its discovery in ''Escherichia coli'' by Larry Taylor was among the first observations of insertion elements in a genome. This discovery opened up the world to an investigation of transposable elements and their effects on a wide variety of organisms. While Mu was specifically involved in several distinct areas of research (including ''E. coli'', maize, and HIV), the wider implications of transposition and insertion transformed the entire field of genetics. Anatomy Phage Mu is nonenveloped, with a head and a tail. The head has an icosahedral structure of about 54 nm in width. The neck is knob-like, and the tail is contractile with a base plate and six short terminal fibers. The genome has been fully sequenced and consists of 36,717 nucleotides, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RecBCD
Exodeoxyribonuclease V (EC 3.1.11.5, RecBCD, Exonuclease V, ''Escherichia coli'' exonuclease V, ''E. coli'' exonuclease V, gene recBC endoenzyme, RecBC deoxyribonuclease, gene recBC DNase, gene recBCD enzymes) is an enzyme of ''E. coli'' that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA. It catalyses exonucleolytic cleavage (in the presence of ATP) in either 5′- to 3′- or 3′- to 5′-direction to yield 5′-phosphooligonucleotides. Structure The enzyme complex is composed of three different subunits called RecB, RecC, and RecD and hence the complex is named RecBCD (Figure 1). Before the discovery of the ''recD'' gene, the enzyme was known as “RecBC.” Each subunit is encod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Concatemer
A concatemer is a long continuous DNA molecule that contains multiple copies of the same DNA sequence linked in series. These polymeric molecules are usually copies of an entire genome linked end to end and separated by ''cos'' sites (a protein binding nucleotide sequence that occurs once in each copy of the genome). Concatemers are frequently the result of rolling circle replication, and may be seen in the late stage of bacterial infection by phages. As an example, if the genes in the phage DNA are arranged ABC, then in a concatemer the genes would be ABCABCABCABC and so on (assuming synthesis was initiated between genes C and A). They are further broken by ribozymes. During active infection, some species of viruses have been shown to replicate their genetic material via the formation of concatemers. In the case of ''human herpesvirus-6'', its entire genome is made over and over on a single strand. These long concatemers are subsequently cleaved between the pac-1 and pac-2 region ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enterobacteria Phage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains (or isolates), Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6. Use in research Dating back to the 1940s and continuing today, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phage's genetic information is made up of around 300 genes. Coincident with their complexity, T-even viruses were found to have the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |