|
Oxaloacetate Decarboxylase
Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate. It is categorized under . Oxaloacetate decarboxylase activity in a given organism may be due to activity of malic enzyme, pyruvate kinase, malate dehydrogenase, pyruvate carboxylase and PEP carboxykinase or the activity of "real" oxaloacetate decarboxylases. The latter enzymes catalyze the irreversible decarboxylation of oxaloacetate and can be classified into (i) the divalent cation-dependent oxaloacetate decarboxylases and (ii) the membrane-bound sodium-dependent and biotin-containing oxaloacetate decarboxylases from enterobacteria. Kinetic Properties An oxaloacetate decarboxylase from the family of divalent cation dependent decarboxylases was isolated from Corynebacterium glutamicum in 1995 by Jetten et al. This enzyme selectively catalyzed the decarboxylation of oxaloacetate to pyruvate and CO2 with a Km of 2.1mM, Vmax of 158 umol, and kcat of 311 s^-1. Mn2+ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
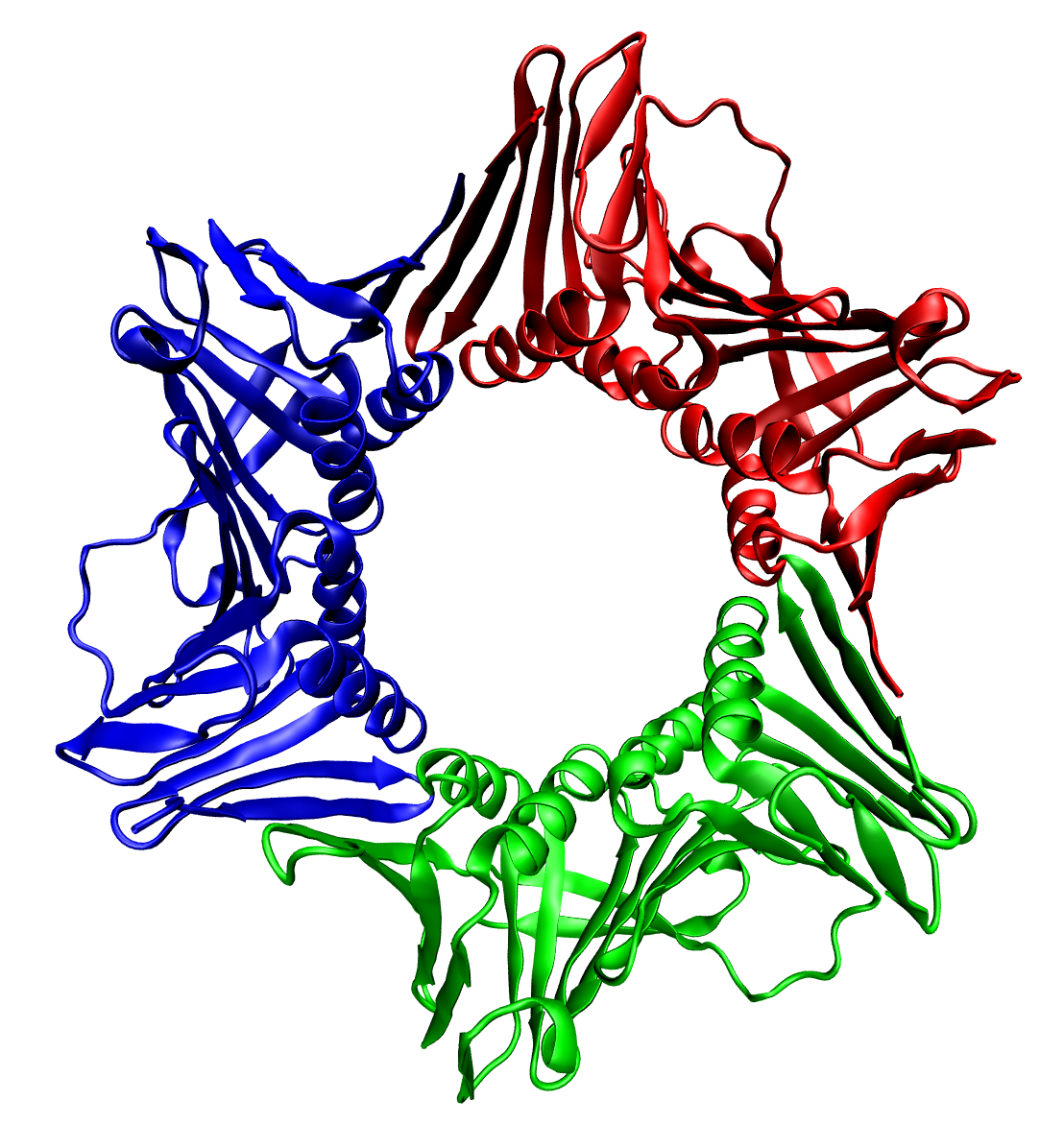
Crystal Structure Of Oxaloacetate Decarboxylase Na+ Pump
A crystal or crystalline solid is a solid material whose constituents (such as atoms, molecules, or ions) are arranged in a highly ordered microscopic structure, forming a crystal lattice that extends in all directions. In addition, macroscopic single crystals are usually identifiable by their geometrical shape, consisting of flat face (geometry), faces with specific, characteristic orientations. The scientific study of crystals and crystal formation is known as crystallography. The process of crystal formation via mechanisms of crystal growth is called crystallization or solidification. The word ''crystal'' derives from the Ancient Greek word (), meaning both "ice" and "Quartz#Varieties (according to color), rock crystal", from (), "icy cold, frost". Examples of large crystals include snowflakes, diamonds, and table salt. Most inorganic solids are not crystals but polycrystals, i.e. many microscopic crystals fused together into a single solid. Polycrystals include most m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Veillonella Parvula
''Veillonella parvula'' is a strictly anaerobic, Gram-negative, coccus-shaped bacterium in the genus ''Veillonella''. It is a normal part of the oral flora but can be associated with diseases such as periodontitis and dental caries as well as various systemic infections, including meningitis and osteomyelitis. It has also been isolated from women with bacterial vaginosis and has been associated with hypertension together with ''Campylobacter rectus'' and ''Prevotella melaninogenica''. ''V. parvula'' is unable to feed on carbohydrates, but can feed on lactate provided by ''Streptococcus'' species also found in the oral cavity. Specifically, ''Streptococcus mutans'' and ''V. parvula'' can form multispecies biofilms that lead to a lower susceptibility to antimicrobial treatments, resulting in periodontitis and dental carries. Antimicrobial susceptibility ''V. parvula'' bacteria are susceptible to and typically treated with metronidazole and penicillin in clinical reports. Other an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integral Membrane Proteins
An integral, or intrinsic, membrane protein (IMP) is a type of membrane protein that is permanently attached to the biological membrane. All ''transmembrane proteins'' are IMPs, but not all IMPs are transmembrane proteins. IMPs comprise a significant fraction of the proteins encoded in an organism's genome. Proteins that cross the membrane are surrounded by annular lipids, which are defined as lipids that are in direct contact with a membrane protein. Such proteins can only be separated from the membranes by using detergents, nonpolar solvents, or sometimes denaturing agents. Structure Three-dimensional structures of ~160 different integral membrane proteins have been determined at atomic resolution by X-ray crystallography or nuclear magnetic resonance spectroscopy. They are challenging subjects for study owing to the difficulties associated with extraction and crystallization. In addition, structures of many water-soluble protein domains of IMPs are available in the Prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally-occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Trimer
In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein. Porins usually arrange themselves in membranes as trimers. Bacteriophage T4 tail fiber Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation. The distal portion of each of the bacteriophage T4 tail fibers is encoded by ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carboxybiotin Decarboxylase
Carboxybiotin decarboxylase (, ''MadB'', ''carboxybiotin protein decarboxylase'') is an enzyme with systematic name ''carboxybiotinyl-(protein) carboxy-lyase''. This enzyme catalyses the following chemical reaction : a carboxybiotinyl- rotein+ ''n'' Na+in + H+out \rightleftharpoons CO2 + a biotinyl- rotein+ ''n'' Na+out (''n'' = 1--2) This enzyme is an integral membrane protein MadB from the anaerobic bacterium '' Malonomonas rubra''. Nomenclature This enzyme was previously classified as . References External links * {{Portal bar, Biology, border=no EC 4.3.99 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium Ions
Sodium is a chemical element with the symbol Na (from Latin ''natrium'') and atomic number 11. It is a soft, silvery-white, highly reactive metal. Sodium is an alkali metal, being in group 1 of the periodic table. Its only stable isotope is 23Na. The free metal does not occur in nature, and must be prepared from compounds. Sodium is the sixth most abundant element in the Earth's crust and exists in numerous minerals such as feldspars, sodalite, and halite (NaCl). Many salts of sodium are highly water-soluble: sodium ions have been leached by the action of water from the Earth's minerals over eons, and thus sodium and chlorine are the most common dissolved elements by weight in the oceans. Sodium was first isolated by Humphry Davy in 1807 by the electrolysis of sodium hydroxide. Among many other useful sodium compounds, sodium hydroxide ( lye) is used in soap manufacture, and sodium chloride ( edible salt) is a de-icing agent and a nutrient for animals ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gibbs Free Energy
In thermodynamics, the Gibbs free energy (or Gibbs energy; symbol G) is a thermodynamic potential that can be used to calculate the maximum amount of work that may be performed by a thermodynamically closed system at constant temperature and pressure. It also provides a necessary condition for processes such as chemical reactions that may occur under these conditions. The Gibbs free energy change , measured in joules in SI) is the ''maximum'' amount of non-expansion work that can be extracted from a closed system (one that can exchange heat and work with its surroundings, but not matter) at fixed temperature and pressure. This maximum can be attained only in a completely reversible process. When a system transforms reversibly from an initial state to a final state under these conditions, the decrease in Gibbs free energy equals the work done by the system to its surroundings, minus the work of the pressure forces. The Gibbs energy is the thermodynamic potential that is minim ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Na+ Pump
NA, N.A., Na, nA or n/a may refer to: Chemistry and physics * Sodium, symbol Na, a chemical element * Avogadro constant (''N''A) * Nucleophilic addition, a type of reaction in organic chemistry * Numerical aperture, a number that characterizes a range of angles in an optical system * nA, the symbol for nanoampere * Naturally aspirated engine Biology and medicine * Na (tree) or ''Mesua ferrea'', a species of tree native to Sri Lanka * Neuroacanthocytosis, a neurological condition * '' Nomina Anatomica'', a former international standard for human anatomical nomenclature * Noradrenaline, a hormone * Nucleic acid analogue, compounds analogous to naturally occurring RNA and DNA Places Current * Namibia (ISO country code) * Naples (car number plate code: NA), Italy * North America, a continent * North Africa, a subcontinent Historical * Netherlands Antilles (former international vehicle registration code: NA) * Na (Chinese state), a small state of the Chinese Zhou dynasty from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Diphosphate
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenine attaches to the 1’ carbon. ADP can be interconverted to adenosine triphosphate (ATP) and adenosine monophosphate (AMP). ATP contains one more phosphate group than does ADP. AMP contains one fewer phosphate group. Energy transfer used by all living things is a result of dephosphorylation of ATP by enzymes known as ATPases. The cleavage of a phosphate group from ATP results in the coupling of energy to metabolic reactions and a by-product of ADP. ATP is continually reformed from lower-energy species ADP and AMP. The biosynthesis of ATP is achieved through ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |