|
NrrF RNA
NrrF is a non-coding RNA which is regulated by the Ferric uptake regulator (Fur) protein in bacteria. This non-coding RNA was identified in ''Neisseria meningitidis'' and is involved in iron regulation of the succinate dehydrogenase genes sdhA and sdhC. NrrF acts as an antisense RNA and is complementary to the junction between the second and third genes of the sdh operon (sdhD and sdhA). Secondary structure predictions have indicated that this interaction occurs in a single stranded loop region of the NrrF RNA. Under low iron concentration NrrF is present at a high concentration and forms a duplex with the transcript in Hfq dependent manner. The RNA chaperone Hfq acts to enhance binding of NrrF or stabilizes the NrrF/sdh transcript duplex. Binding of NrrF results in down regulation of the sdhCDAB mRNA transcript results in a Fur-dependent positive regulation of succinate dehydrogenase. Another NrrF RNA target is mRNA ''petABC'', coding for cytochrome ''bc1'' ( an essential compo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
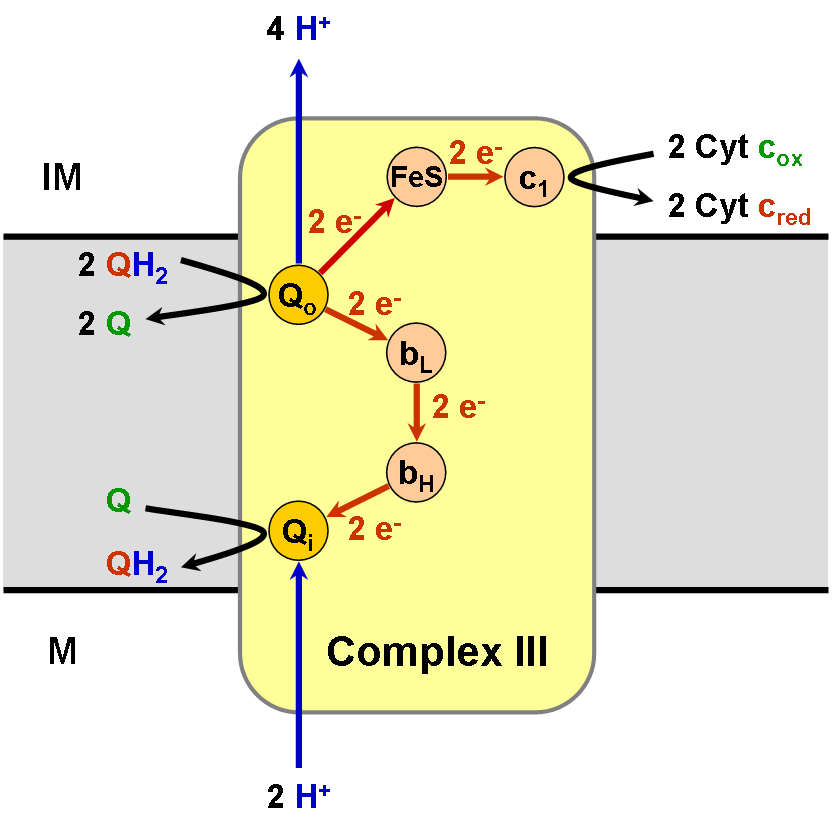
Coenzyme Q – Cytochrome C Reductase
The coenzyme Q : cytochrome ''c'' – oxidoreductase, sometimes called the cytochrome ''bc''1 complex, and at other times complex III, is the third complex in the electron transport chain (), playing a critical role in biochemical generation of ATP (oxidative phosphorylation). Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial (cytochrome b) and the nuclear genomes (all other subunits). Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits (cytochrome B, cytochrome C1, Rieske protein), 2 core proteins and 6 low-molecular weight proteins. Ubiquinol—cytochrome-c reductase catalyzes the chemical reaction :QH2 + 2 ferricytochrome c \rightleftharpoons Q + 2 ferrocytochrome c + 2 H+ Thus, the two substrates of this enzyme ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neisseria RNA Thermometers
RNA thermometers (RNATs) regulate gene expression in response to temperature, allowing pathogens such as '' Neisseria meningitidis'' to switch on silent genes after entering the host organism. However the temperature for expression of Neisseria virulence-associated traits is 42 °C while other bacterial pathogen RNATs require 37 °C. This is probably because ''N. meningitidis'' is an obligate commensal of the human nasopharynx and becomes pathogenic during inflammation due to viral infection. Three independent RNA thermosensors were identified in the 5′UTRs of genes needed for: capsule biosynthesis (cssA), the expression of factor H binding protein (fHbp) and sialylation of lipopolysaccharide, which is essential for bacterial resistance against immune killing (lst). The very different nucleotide sequence and predicted inhibitory structures of the three RNATs indicate that they have evolved independently. Further study of molecular mechanisms of CssA RNAT show ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neisseria Sigma-E SRNA
Neisseria sigma-E sRNA (NSE sRNA) is a non-coding RNA found in the bacterial genus Neisseria, including the two pathogens N. meningitidis and N. gonorrhoeae. The RNA was discovered in a screen for genes differentiated by high expression of the Sigma factor, sigma E. Seven genes were predicted to be regulated by NSE sRNA, including fur, nadC and a putative TetR family transcriptional regulator, through base-pairing interactions with the 5' UTR. The sRNA transcript is terminated by a rho independent terminator. See also: * NrrF sRNA * Neisseria RNA thermometers * Neisseria sibling sRNAs NmsR/RcoF NmsRA and NmsRB (Neisseria metabolic switch regulator), RcoF1 and RcoF2 (RNA regulating colonization factor) as well as NgncR_162 and NgncR_163 (Neisseria gonorrhoeae non-coding RNA) are all names of neisserial sibling small regulatory RNAs descri ... References {{reflist RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aggregatibacter Iron-regulated SRNA
Four non-coding small RNAs containing a Fur box-like ( ferric uptake regulator) sequence were identified by bioinformatics analysis in ''Aggregatibacter actinomycetemcomitans'' ''HK1651'' called JA01-JA04. The transcription of sRNAs was confirmed by Northern blot. Fur binding was demonstrated to each sRNA promoter, and that transcription of the sRNAs was decreased in presence of iron and increased by iron limitation. JA03 may have the ability to regulate biofilm formation. JA01 is conserved only among ''A. actinomycetemcomitans''. JA02 is present in both ''A. actinomycetemcomitans and P. multocida. J''A 03 and JA04 are most widely conserved and have orthologues across many ''Pasteurellaceae''. HrrF RNA HrrF RNA (''Haemophilus'' regulatory RNA responsive to iron Fe) is a small non-coding RNA involved in iron homeostasis in ''Haemophilus'' species. Orthologues exist only among other '' Pasteurellacae.'' Iron- regulated sRNAs JA01- JA04 were ident ... is another Fur-regulated sRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HrrF RNA
HrrF RNA (''Haemophilus'' regulatory RNA responsive to iron Fe) is a small non-coding RNA involved in iron homeostasis in ''Haemophilus'' species. Orthologues exist only among other '' Pasteurellacae.'' Iron- regulated sRNAs JA01- JA04 were identified in related ''Aggregatibacte''r. It is an analog to PrrF and RyhB RNAs. HrrF is maximally expressed when iron levels are low. Ferric uptake regulator (Fur) binds upstream of the ''hrrF'' promoter. HrrF stability is not dependent on the RNA chaperone Hfq. RNA-seq has shown that HrrF targets are mRNAs of genes whose products are involved in molybdate uptake, deoxyribonucleotide synthesis, and amino acid synthesis. See also * NrrF RNA NrrF is a non-coding RNA which is regulated by the Ferric uptake regulator (Fur) protein in bacteria. This non-coding RNA was identified in ''Neisseria meningitidis'' and is involved in iron regulation of the succinate dehydrogenase genes sdhA an ... References Non-coding RNA {{mole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PrrF RNA
The PrrF RNAs are small non-coding RNAs involved in iron homeostasis and are encoded by all ''Pseudomonas'' species. The PrrF RNAs are analogs of the RyhB RNA, which is encoded by enteric bacteria. Expression of the PrrF RNAs is repressed by the ferric uptake regulator (Fur) when cells are grown in iron-replete conditions. Under iron limitation, the PrrF RNAs are expressed and act to negatively regulate several genes encoding iron-containing proteins, including SodB and succinate dehydrogenase. As such, PrrF regulation "spares" iron when this nutrient becomes scarce. PrrF and virulence In ''Pseudomonas aeruginosa'', the PrrF RNAs are required for the production of the Pseudomonas quinolone signal (PQS), a quorum sensing molecule that activates the expression of several virulence genes. The phenomenon is mediated by PrrF repressing the expression of enzymes that degrade anthranilic acid, which is the precursor for PQS synthesis. Due to this regulatory link with PQS, the PrrF RNAs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RyhB RNA
RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur ( Ferric uptake regulator). Discovery The gene was independently identified in two screens, named RyhB by Wassarman ''et al''. and called SraI by Argaman ''et al''. and was found to be expressed only in stationary phase. Function and regulation RyhB RNA levels are inversely correlated with mRNA levels for the ''sdhCDAB'' operon, encoding succinate dehydrogenase, as well as five other genes previously shown to be positively regulated by Fur by an unknown mechanism. These include two other genes encoding enzymes in the tricarboxylic acid cycle, ''acnA'' and ''fumA'', two ferritin genes, ''ftnA'' and ''bfr'', and a gene for superoxide dismutase, ''sodB''. A number of other genes have been predicted computationally and verified as targets by microarray analysis: ''napF'', ''sodA'', ''cysE ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chaperone (protein)
In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to assist large proteins in proper protein folding during or after synthesis, and after partial denaturation. Chaperones are also involved in the translocation of proteins for proteolysis. The first molecular chaperones discovered were a type of assembly chaperones which assist in the assembly of nucleosomes from folded histones and DNA. One major function of molecular chaperones is to prevent the aggregation of misfolded proteins, thus many chaperone proteins are classified as heat shock proteins, as the tendency for protein aggregation is increased by heat stress. The majority of molecular chaperones do not convey any steric information for protein folding, and instead assist in protein folding by binding to and stabilizing folding interme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antisense RNA
Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs (which occur naturally) have been found in both prokaryotes and eukaryotes, and can be classified into short (200 nucleotides) non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications. Discovery and history in drug development Some of the earliest asRNAs were discovered while investigating functional proteins. An example was micF asRNA. While characterizing the outer membrane porin in ''E.coli'', some of the promoter clones observed were capable of repressing the expression of other membrane porin s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Succinate Dehydrogenase
Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential. In step 6 of the citric acid cycle, SQR catalyzes the oxidation of succinate to fumarate with the reduction of ubiquinone to ubiquinol. This occurs in the inner mitochondrial membrane by coupling the two reactions together. Structure Subunits Mitochondrial and many bacterial SQRs are composed of four structurally different subunits: two hydrophilic and two hydrophobic. The first two subunits, a flavoprotein (SdhA) and an iron-sulfur protein (SdhB), form a hydrophilic head where enzymatic activity of the complex takes place. Sdh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



_.png)