|
Non-homologous End-joining Factor 1
Non-homologous end-joining factor 1 (NHEJ1), also known as Cernunnos or XRCC4-like factor (XLF), is a protein that in humans is encoded by the NHEJ1 gene. XLF was originally discovered as the protein mutated in five patients with growth retardation, microcephaly, and immunodeficiency. The protein is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Patients with XLF mutations also have immunodeficiency due to a defect in V(D)J recombination, which uses NHEJ to generate diversity in the antibody repertoire of the immune system. XLF interacts with DNA ligase IV and XRCC4 and is thought to be involved in the end-bridging or ligation steps of NHEJ. The yeast (''Saccharomyces cerevisiae'') homolog of XLF is Nej1. Phenotypes In contrast to the profound immunodeficiency phenotype of XLF deletion in humans, deletion of XLF alone has a mild phenotype in mice. However, combining a deletion of XLF with deletion of the ATM kinase causes a synthetic defect i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malignant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
V(D)J Recombination
V(D)J recombination is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire of antibodies/immunoglobulins and T cell receptors (TCRs) found in B cells and T cells, respectively. The process is a defining feature of the adaptive immune system. V(D)J recombination in mammals occurs in the primary lymphoid organs (bone marrow for B cells and thymus for T cells) and in a nearly random fashion rearranges variable (V), joining (J), and in some cases, diversity (D) gene segments. The process ultimately results in novel amino acid sequences in the antigen-binding regions of immunoglobulins and TCRs that allow for the recognition of antigens from nearly all pathogens including bacteria, viruses, parasites, and worms as well as "altered self cells" as seen in cancer. The recognition can also be allergic in nature (''e.g.'' to pollen or other allergens) or may match ho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LIG4
DNA ligase 4 is an enzyme that in humans is encoded by the LIG4 gene. Function The protein encoded by this gene is an ATP-dependent DNA ligase that joins double-strand breaks during the non-homologous end joining pathway of double-strand break repair. It is also essential for V(D)J recombination. Lig4 forms a complex with XRCC4, and further interacts with the DNA-dependent protein kinase (DNA-PK) and XLF/Cernunnos, which are also required for NHEJ. The crystal structure of the Lig4/XRCC4 complex has been resolved. Defects in this gene are the cause of LIG4 syndrome. The yeast homolog of Lig4 is Dnl4. LIG4 Syndrome In humans, deficiency of DNA ligase 4 results in a clinical condition known as LIG4 syndrome. This syndrome is characterized by cellular radiation sensitivity, growth retardation, developmental delay, microcephaly, facial dysmorphisms, increased disposition to leukemia, variable degrees of immunodeficiency and reduced number of blood cells. Haematopoietic st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
XRCC4
DNA repair protein XRCC4 also known as X-ray repair cross-complementing protein 4 or XRCC4 is a protein that in humans is encoded by the XRCC4 gene. In addition to humans, the XRCC4 protein is also expressed in many other metazoans, fungi and in plants. The X-ray repair cross-complementing protein 4 is one of several core proteins involved in the non-homologous end joining (NHEJ) pathway to repair DNA double strand breaks (DSBs). NHEJ requires two main components to achieve successful completion. The first component is the cooperative binding and phosphorylation of artemis by the catalytic subunit of the DNA-dependent protein kinase ( DNA-PKcs). Artemis cleaves the ends of damaged DNA to prepare it for ligation. The second component involves the bridging of DNA to DNA Ligase IV ( LigIV), by XRCC4, with the aid of Cernunnos-XLF. DNA-PKcs and XRCC4 are anchored to Ku70 / Ku80 heterodimer, which are bound to the DNA ends. Since XRCC4 is the key protein that enables interacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular biology, molecular and cell biology, much like ''Escherichia coli'' as the model bacteria, bacterium. It is the microorganism behind the most common type of fermentation (biochemistry), fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 micrometre, μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their Homology (biology), homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley body, Berkeley bo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
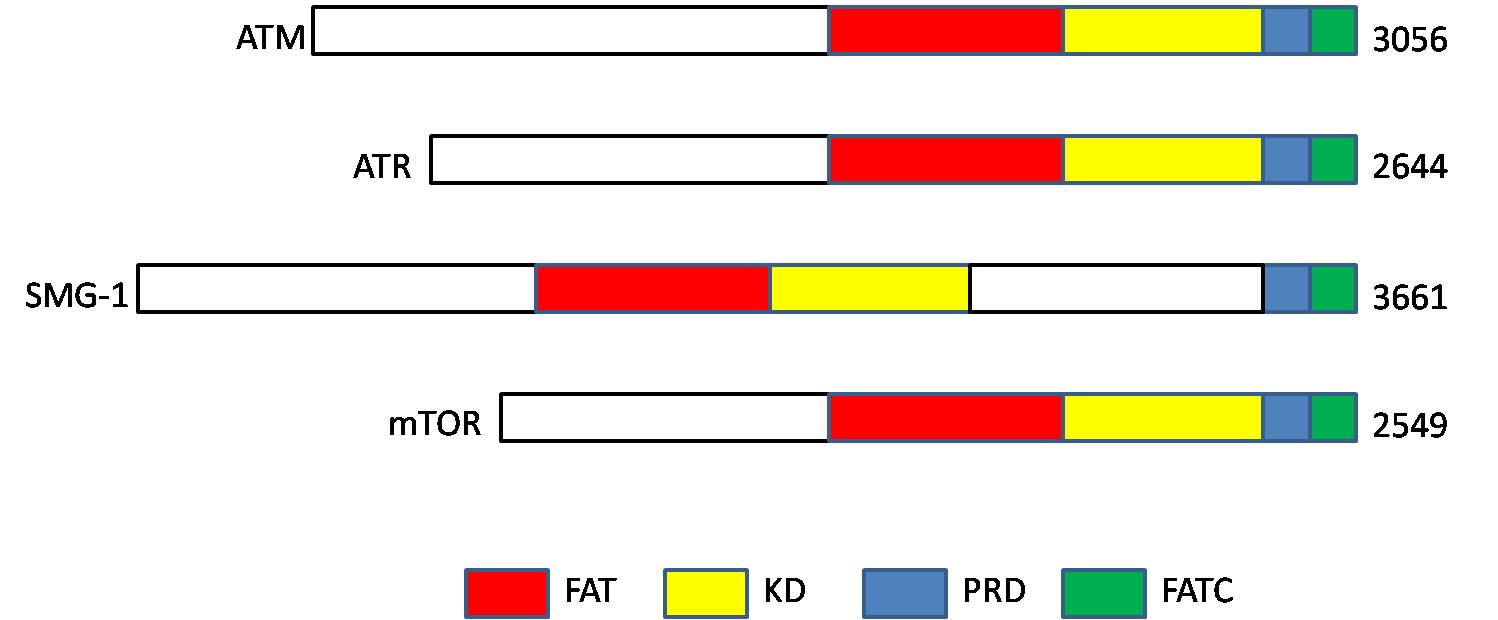
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during DNA replication, replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has diffe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ku (protein)
Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer (two copies of the same protein bound to each other). Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation. The Ku70 and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |