|
Nitroreductase
Nitroreductases are a family of evolutionarily related proteins involved in the reduction of nitrogen-containing compounds, including those containing the nitro functional group. Members of this family utilise flavin mononucleotide as a cofactor and are often found to be homodimers. Members of this family include oxygen-insensitive NAD(P)H nitroreductase (flavin mononucleotide-dependent nitroreductase) (6,7-dihydropteridine reductase) () and NADH dehydrogenase (). A number of these proteins are described as oxidoreductases. They are primarily found in bacterial lineages though a number of eukaryotic homologs have been identified: ''C. elegans'' , ''D. melanogaster'' , , mouse and human . This protein is not found in photosynthetic eukaryotes. The sequences containing this entry in photosynthetic organisms are possible false positives. The nitroreductase of ''Enterobacter cloacae'' was identified by Bryant and Deluca in a strain isolated from a munitions facility, on the basis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with Family (biology), family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar protein structure, three-dimensional structures, functions, and significant Sequence homology, sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitro Compound
In organic chemistry, nitro compounds are organic compounds that contain one or more nitro functional groups (). The nitro group is one of the most common explosophores (functional group that makes a compound explosive) used globally. The nitro group is also strongly electron-withdrawing. Because of this property, bonds alpha (adjacent) to the nitro group can be acidic. For similar reasons, the presence of nitro groups in aromatic compounds retards electrophilic aromatic substitution but facilitates nucleophilic aromatic substitution. Nitro groups are rarely found in nature. They are almost invariably produced by nitration reactions starting with nitric acid. Synthesis Preparation of aromatic nitro compounds Aromatic nitro compounds are typically synthesized by nitration. Nitration is achieved using a mixture of nitric acid and sulfuric acid, which produce the nitronium ion (), which is the electrophile: + The nitration product produced on the la ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flavin Mononucleotide
Flavin mononucleotide (FMN), or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin (vitamin B2) by the enzyme riboflavin kinase and functions as the prosthetic group of various oxidoreductases, including NADH dehydrogenase, as well as cofactor in biological blue-light photo receptors. During the catalytic cycle, a reversible interconversion of the oxidized (FMN), semiquinone (FMNH•), and reduced (FMNH2) forms occurs in the various oxidoreductases. FMN is a stronger oxidizing agent than NAD and is particularly useful because it can take part in both one- and two-electron transfers. In its role as blue-light photo receptor, (oxidized) FMN stands out from the 'conventional' photo receptors as the signaling state and not an E/Z isomerization. It is the principal form in which riboflavin is found in cells and tissues. It requires more energy to produce, but is more soluble than riboflavin. In cells, FMN occurs freely circulating but also in several covalently b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
6,7-dihydropteridine Reductase
In enzymology, 6,7-dihydropteridine reductase (, also Dihydrobiopterin reductase) is an enzyme that catalyzes the chemical reaction : 5,6,7,8-tetrahydropteridine + NAD(P)+ \rightleftharpoons 6,7-dihydropteridine + NAD(P)H + H+ The four substrates for this enzyme are a 6,7-dihydropteridine (dihydrobiopterin), NADH, NADPH, and H+ and its three products are 5,6,7,8-tetrahydropteridine (tetrahydrobiopterin), NAD+, and NADP+ This enzyme participates in folate biosynthesis. In the human genome, the enzyme is encoded by the QDPR gene. Nomenclature This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donors with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is 5,6,7,8-tetrahydropteridine:NAD(P)+ oxidoreductase. Other names in common use include 6,7-dihydropteridine:NAD(P)H oxidoreductase, DHPR, NAD(P)H:6,7-dihydropteridine oxidoreductase, NADH-dihydropteridine reductase, NADPH-dihydropteridine reductase, NADPH- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADH Dehydrogenase
NADH dehydrogenase is an enzyme that converts nicotinamide adenine dinucleotide (NAD) from its reduced form (NADH) to its oxidized form (NAD+). Members of the NADH dehydrogenase family and analogues are commonly systematically named using the format ''NADH:acceptor oxidoreductase''. The chemical reaction these enzymes catalyze are generally represented with the follow equation; : NADH + H+ + acceptor NAD+ + reduced acceptor NADH dehydrogenase is a flavoprotein that contains iron-sulfur centers. NADH dehydrogenase is used in the electron transport chain for generation of ATP. The EC term NADH dehydrogenase (quinone) (EC 1.6.5.11) is defined for NADH dehydrogenases that use a quinone (excluding ubiquinone) as the acceptor. The EC term NADH dehydrogenase (ubiquinone) Respiratory complex I, (also known as NADH:ubiquinone oxidoreductase, Type I NADH dehydrogenase and mitochondrial complex I) is the first large protein complex of the respiratory chains of many organisms from bac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caenorhabditis Elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (rod-like) and Latin ''elegans'' (elegant). In 1900, Maupas initially named it '' Rhabditides elegans.'' Osche placed it in the subgenus ''Caenorhabditis'' in 1952, and in 1955, Dougherty raised ''Caenorhabditis'' to the status of genus. ''C. elegans'' is an unsegmented pseudocoelomate and lacks respiratory or circulatory systems. Most of these nematodes are hermaphrodites and a few are males. Males have specialised tails for mating that include spicules. In 1963, Sydney Brenner proposed research into ''C. elegans,'' primarily in the area of neuronal development. In 1974, he began research into the molecular and developmental biology of ''C. elegans'', which has since been extensively used as a model organism. It was the first multicellu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Melanogaster
''Drosophila melanogaster'' is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly" or "pomace fly". Starting with Charles W. Woodworth's 1901 proposal of the use of this species as a model organism, ''D. melanogaster'' continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and life history evolution. As of 2017, five Nobel Prizes have been awarded to drosophilists for their work using the insect. ''D. melanogaster'' is typically used in research owing to its rapid life cycle, relatively simple genetics with only four pairs of chromosomes, and large number of offspring per generation. It was originally an African species, with all non-African lineages having a common origin. Its geographic range includes all continents, including islands. ''D. melanogaster'' is a common pest in homes, restaurants, and othe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
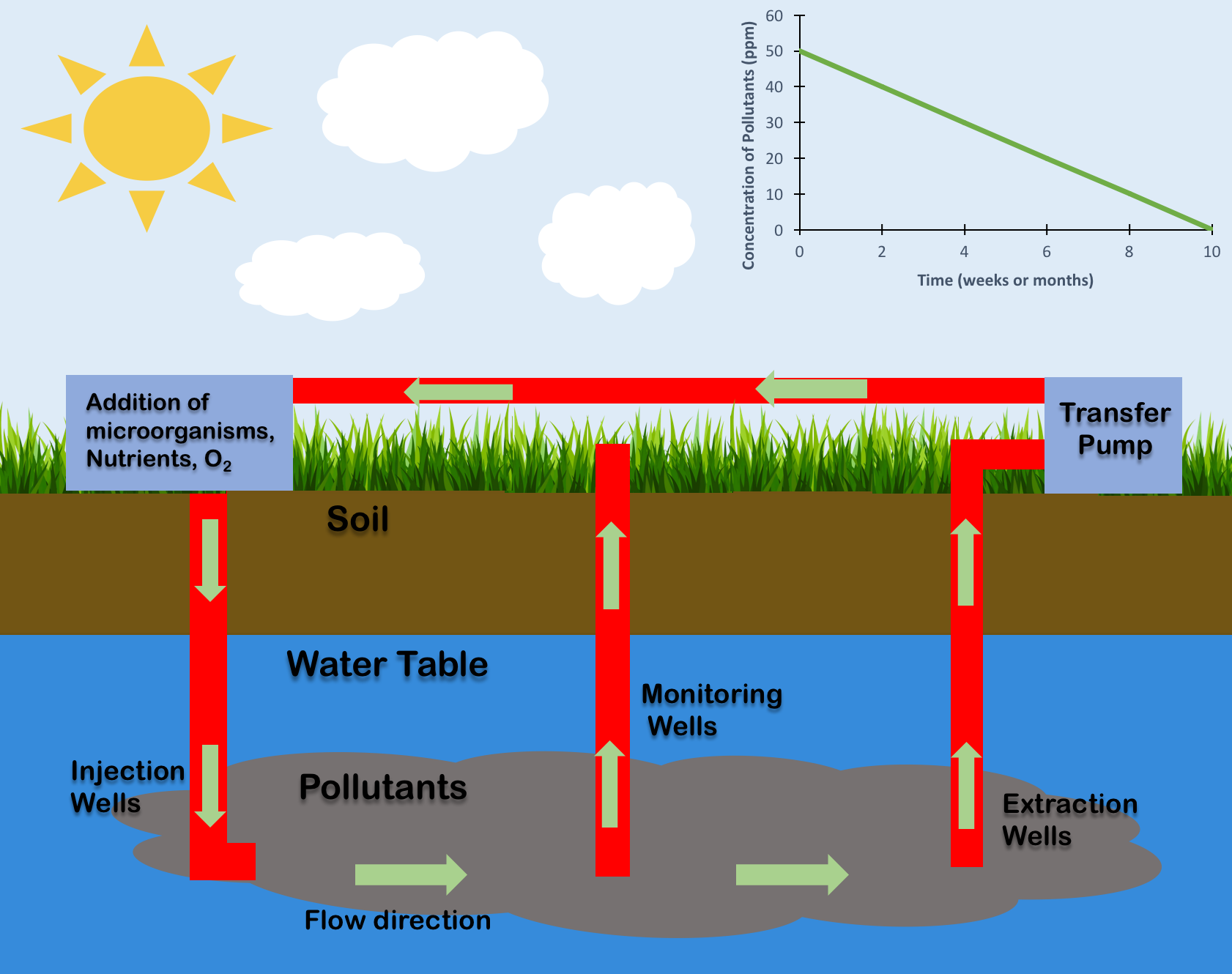
Bioremediation
Bioremediation broadly refers to any process wherein a biological system (typically bacteria, microalgae, fungi, and plants), living or dead, is employed for removing environmental pollutants from air, water, soil, flue gasses, industrial effluents etc., in natural or artificial settings. The natural ability of organisms to adsorb, accumulate, and degrade common and emerging pollutants has attracted the use of biological resources in treatment of contaminated environment. In comparison to conventional physicochemical treatment methods bioremediation may offer considerable advantages as it aims to be sustainable, eco-friendly, cheap, and scalable. Most bioremediation is inadvertent, involving native organisms. Research on bioremediation is heavily focused on stimulating the process by inoculation of a polluted site with organisms or supplying nutrients to promote the growth. In principle, bioremediation could be used to reduce the impact of byproducts created from anthropogenic acti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cob(II)yrinic Acid A,c-diamide Reductase
In enzymology, a cob(II)yrinic acid a,c-diamide reductase () is an enzyme that catalyzes Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ... the chemical reaction 2 cob(I)yrinic acid a,c-diamide + FMN + 3 H+ \rightleftharpoons 2 cob(II)yrinic acid a,c-diamide + FMNH2 The three substrates of this enzyme are cob(I)yrinic acid a,c-diamide, flavin mononucleotide, and H+; its two products are cob(II)yrinic acid a,c-diamide and FMNH2. Classification This enzyme belongs to the family of oxidoreductases, specifically those oxidizing metal ion with a flavin as acceptor. Nomenclature The systematic name of this enzyme class is cob(I)yrinic acid-a,c-diamide:FMN oxidoreductase. This enzyme is also called CobR and cob(II)yrinic acid-a,c-diamide:FMN oxidoreductase (incorrect). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domains
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer after ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |