|
Myb Domain
Myb genes are part of a large gene family of transcription factors found in animals and plants. In humans, it includes Myb proto-oncogene like 1 and Myb-related protein B in addition to MYB proper. Members of the extended SANT/Myb family also include the SANT domain and other similar all-helical homeobox-like domains. Function Viral The Myb gene family is named after the eponymous gene in Avian myeloblastosis virus. The viral Myb (v-Myb, ) recognizes the sequence 5'-YAACKG-3'. It causes myeloblastosis (myeloid leukemia) in chickens. Compared to the normal animal cellular Myb (c-myb), v-myb contains deletions in the C-terminal regulatory domain, leading to aberrant activation of other oncogenes. Animals Myb proto-oncogene protein is a member of the MYB (myeloblastosis) family of transcription factors. The protein contains three domains, an N-terminal DNA-binding domain, a central transcriptional activation domain and a C-terminal domain involved in transcriptional repre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flavan-4-ol
The flavan-4-ols (3-deoxyflavonoids) are flavone-derived alcohols and a family of flavonoids. Flavan-4-ols are colorless precursor compounds that polymerize to form red phlobaphene pigments. They can be found in the sorghum. Glycosides (abacopterins A, B, C and D together with triphyllin A and 6,8-dimethyl-7-hydroxy-4‘-methoxyanthocyanidin-5-O-β-d-glucopyranoside) can be isolated from a methanol extract of the rhizomes of ''Abacopteris penangiana''. Known flavan-4-ols * Apiforol * Luteoforol Metabolism Flavanone 4-reductase is an enzyme that uses (2''S'')-flavan-4-ol and NADP+ to produce (2''S'')-flavanone The flavanones, a type of flavonoids, are various aromatic, colorless ketones derived from flavone that often occur in plants as glycosides. List of flavanones * Blumeatin * Butin * Eriodictyol * Hesperetin * Hesperidin * Homoeriodictyol * Iso ..., NADPH, and H+. Spectral data These compounds have absorption maxima of 564 nm. References {{flavan-4ol [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EZH1
#REDIRECT EZH1 #REDIRECT EZH1 {{R from other capitalization ... {{R from other capitalization ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anthocyanin
Anthocyanins (), also called anthocyans, are water-soluble vacuolar pigments that, depending on their pH, may appear red, purple, blue, or black. In 1835, the German pharmacist Ludwig Clamor Marquart gave the name Anthokyan to a chemical compound that gives flowers a blue color for the first time in his treatise "''Die Farben der Blüthen''". Food plants rich in anthocyanins include the blueberry, raspberry, black rice, and black soybean, among many others that are red, blue, purple, or black. Some of the colors of autumn leaves are derived from anthocyanins. Anthocyanins belong to a parent class of molecules called flavonoids synthesized via the phenylpropanoid pathway. They occur in all tissues of higher plants, including leaves, stems, roots, flowers, and fruits. Anthocyanins are derived from anthocyanidins by adding sugars. They are odorless and moderately astringent. Although approved as food and beverage colorant in the European Union, anthocyanins are not approved ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3-deoxyflavonoids
The flavan-4-ols (3-deoxyflavonoids) are flavone-derived alcohols and a family of flavonoids. Flavan-4-ols are colorless precursor compounds that polymerize to form red phlobaphene pigments. They can be found in the sorghum. Glycosides (abacopterins A, B, C and D together with triphyllin A and 6,8-dimethyl-7-hydroxy-4‘-methoxyanthocyanidin-5-O-β-d-glucopyranoside) can be isolated from a methanol extract of the rhizomes of ''Abacopteris penangiana''. Known flavan-4-ols * Apiforol * Luteoforol Metabolism Flavanone 4-reductase is an enzyme that uses (2''S'')-flavan-4-ol and NADP+ to produce (2''S'')-flavanone The flavanones, a type of flavonoids, are various aromatic, colorless ketones derived from flavone that often occur in plants as glycosides. List of flavanones * Blumeatin * Butin * Eriodictyol * Hesperetin * Hesperidin * Homoeriodictyol * Iso ..., NADPH, and H+. Spectral data These compounds have absorption maxima of 564 nm. References {{flavan-4ol [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydroflavonol Reductase
In enzymology, a dihydrokaempferol 4-reductase () is an enzyme that catalyzes the chemical reaction :cis-3,4-leucopelargonidin + NADP+ \rightleftharpoons (+)-dihydrokaempferol + NADPH + H+ Thus, the two substrates of this enzyme are cis-3,4-leucopelargonidin and NADP+, whereas its 3 products are (+)-dihydrokaempferol, NADPH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is cis-3,4-leucopelargonidin:NADP+ 4-oxidoreductase. Other names in common use include dihydroflavanol 4-reductase (DFR), dihydromyricetin reductase, NADPH-dihydromyricetin reductase, and dihydroquercetin reductase. This enzyme participates in flavonoid biosynthesis. Function Anthocyanidins, common plant pigments, are further reduced by the enzyme dihydroflavonol 4-reductase (DFR) to the corresponding colorless leucoanthocyanidins. DFR uses dihydromyricetin ( am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chalcone Isomerase
In enzymology, a chalcone isomerase () is an enzyme that catalyzes the chemical reaction :a chalcone \rightleftharpoons a flavanone Hence, this enzyme has one substrate, a chalcone, and one product, a flavanone. This enzyme belongs to the family of isomerases, specifically the class of intramolecular lyases. The systematic name of this enzyme class is flavanone lyase (decyclizing). This enzyme is also called chalcone-flavanone isomerase. This enzyme participates in flavonoid biosynthesis. The ''Petunia hybrida'' (Petunia) genome contains two genes coding for very similar enzymes, ChiA and ChiB, but only the first seems to encode a functional chalcone isomerase. Structural studies As of late 2007, 7 structures have been solved for this class of enzymes, with PDB accession codes , , , , , , and . Chalcone isomerase has a core 2-layer alpha/beta structure A structure is an arrangement and organization of interrelated elements in a material object or system, or the object ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
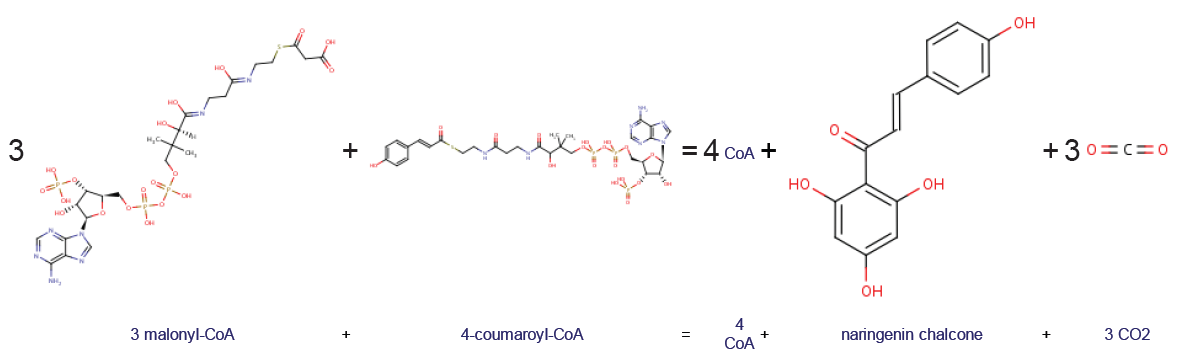
Chalcone Synthase
Chalcone synthase or naringenin-chalcone synthase (CHS) is an enzyme ubiquitous to higher plants and belongs to a family of polyketide synthase enzymes (PKS) known as type III PKS. Type III PKSs are associated with the production of chalcones, a class of organic compounds found mainly in plants as natural defense mechanisms and as synthetic intermediates. CHS was the first type III PKS to be discovered. It is the first committed enzyme in flavonoid biosynthesis. The enzyme catalyzes the conversion of 4-coumaroyl-CoA and malonyl-CoA to naringenin chalcone. Function CHS catalysis serves as the initial step for flavonoid biosynthesis. Flavonoids are important plant secondary metabolites that serve various functions in higher plants. These include pigmentation, UV protection, fertility, antifungal defense and the recruitment of nitrogen-fixing bacteria. CHS is believed to act as a central hub for the enzymes involved in the flavonoid pathway. Studies have shown that these enzyme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptional Suppressor
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dependent on the genetic background in which it appears. Epistatic mutations therefore have different effects on their own than when they occur together. Originally, the term ''epistasis'' specifically meant that the effect of a gene variant is masked by that of a different gene. The concept of ''epistasis'' originated in genetics in 1907 but is now used in biochemistry, computational biology and evolutionary biology. The phenomenon arises due to interactions, either between genes (such as mutations also being needed in regulators of gene expression) or within them (multiple mutations being needed before the gene loses function), leading to non-linear effects. Epistasis has a great influence on the shape of evolutionary landscapes, which le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydroflavonol
The flavanonols (with two "o"s a.k.a. 3-hydroxyflavanone or 2,3-dihydroflavonol) are a class of flavonoids that use the 3-hydroxy-2,3-dihydro-2-phenylchromen-4-one (IUPAC name) backbone. Some examples include: * Taxifolin (or Dihydroquercetin) * Aromadedrin (or Dihydrokaempferol) * Engeletin (or Dihydrokaempferol-3-rhamnoside) Metabolism * Flavanone 3-dioxygenase * Flavonol synthase * Dihydroflavonol 4-reductase Glycosides Glycosides ( chrysandroside A and chrysandroside B) can be found in the roots of ''Gordonia chrysandra''. Xeractinol, a dihydroflavonol C-glucoside, can be isolated from the leaves of ''Paepalanthus argenteus var. argenteus''. Dihydro-flavonol glycosides (astilbin, neoastilbin, isoastilbin, neoisoastilbin, (2''R'', 3''R'')-taxifolin-3'-''O''-β-D-pyranoglucoside) have been identified in the rhizome of ''Smilax glabra ''Smilax glabra'', sarsaparilla, is a plant species in the genus ''Smilax''. It is native to flora of China, China, the Himalayas, and Indo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydroflavonol 4-reductase
In enzymology, a dihydrokaempferol 4-reductase () is an enzyme that catalyzes the chemical reaction :cis-3,4-leucopelargonidin + NADP+ \rightleftharpoons (+)-dihydrokaempferol + NADPH + H+ Thus, the two substrates of this enzyme are cis-3,4-leucopelargonidin and NADP+, whereas its 3 products are (+)-dihydrokaempferol, NADPH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is cis-3,4-leucopelargonidin:NADP+ 4-oxidoreductase. Other names in common use include dihydroflavanol 4-reductase (DFR), dihydromyricetin reductase, NADPH-dihydromyricetin reductase, and dihydroquercetin reductase. This enzyme participates in flavonoid biosynthesis. Function Anthocyanidins, common plant pigments, are further reduced by the enzyme dihydroflavonol 4-reductase (DFR) to the corresponding colorless leucoanthocyanidins. DFR uses dihydromyricetin ( am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptional Activator
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the prom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |