|
Molden
Molden is a general molecular and electronic structure processing program. Major features * Reads output from the ab initio packages GAMESS (US), Gaussian, MOLPRO, PySCF and from semi-empirical packages such as MOPAC, and supports a number of other formats. * Displays molecular orbitals or electron density as contour plots or 3D grid plots and output to a number of graphical formats. * Animates reaction paths and molecular vibrations. * A Z-matrix editor. Molden program has been tested on different platforms, namely Linux, Windows NT, Windows95, Windows2000, WindowsXP, MacOSX, Silicon Graphics IRIX, Sun SunOS and Solaris. Ambfor, the main force field module of Molden, is an external program that can be initialized from Molden. Ambfor admits protein force field Amber and GAFF (General Amber Force Field). Use of Ambfor is automatic when a protein is studied with Molden. The GAFF force field is used only small molecules. Both Amber and GAFF are based on atomic charges. The d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Molecular Graphics Systems
This is a list of notable software systems that are used for visualizing macromolecules. Key The tables below indicate which types of data can be visualized in each system: See also * Biological data visualization * Comparison of nucleic acid simulation software * Comparison of software for molecular mechanics modeling * List of microscopy visualization systems * List of open-source bioinformatics software * Molecular graphics * Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, v ... References External links * A rather detailed, objective, and technical assessment of about 20 tools. * * * {{Chemistry software Chemistry software molecular graphics systems Molecular modelling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MOPAC
MOPAC is a popular computer program used in computational chemistry. It is designed to implement semi-empirical quantum chemistry algorithms, and it runs on Windows, Mac, and Linux. MOPAC2016 is the current version. MOPAC2016 is able to perform calculations on small molecules and enzymes using PM7, PM6, PM3, AM1, MNDO, and RM1. The Sparkle model (for lanthanide chemistry) is also available. Academic users can use this program for free, whereas government and commercial users must purchase the software. MOPAC was largely written by Michael Dewar's research group at the University of Texas at Austin. Its name is derived from ''Molecular Orbital PACkage'', and it is also a pun on the Mopac Expressway that runs around Austin. MOPAC2007 included the new Sparkle/AM1, Sparkle/PM3, RM1 and PM6 models, with an increased emphasis on solid state capabilities. However, it does not have yet MINDO/3, PM5, analytical derivatives, the Tomasi solvation model and intersystem crossing. MOPAC2007 wa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemistry Software For Linux
Chemistry is the scientific study of the properties and behavior of matter. It is a natural science that covers the elements that make up matter to the compounds made of atoms, molecules and ions: their composition, structure, properties, behavior and the changes they undergo during a reaction with other substances. Chemistry also addresses the nature of chemical bonds in chemical compounds. In the scope of its subject, chemistry occupies an intermediate position between physics and biology. It is sometimes called the central science because it provides a foundation for understanding both basic and applied scientific disciplines at a fundamental level. For example, chemistry explains aspects of plant growth (botany), the formation of igneous rocks (geology), how atmospheric ozone is formed and how environmental pollutants are degraded (ecology), the properties of the soil on the moon (cosmochemistry), how medications work (pharmacology), and how to collect DNA evidence a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CMBI
The Center for Molecular and Biomolecular Informatics (CMBI) based in Nijmegen ( Netherlands) does ''in silico'' research on biomolecules. The facility focusses mainly on protein structure and genomes as well as the small molecules and (drug A drug is any chemical substance that causes a change in an organism's physiology or psychology when consumed. Drugs are typically distinguished from food and substances that provide nutritional support. Consumption of drugs can be via insuffla ...s) which they interact with. External links CMBI Bioinformatics organizations Organizations established in 1999 Organisations based in Gelderland 1999 establishments in the Netherlands Nijmegen References {{reflist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also *Car–Parrinello molecular dynamics *Comparison of force-field implementations *Comparison of nucleic acid simulation software *List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling *List of software for nanostructures modeling *Molecular design software *Molecular dynamics *Molecular modeling on GPUs *Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, v ... Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Z-matrix (chemistry)
In chemistry, the Z-matrix is a way to represent a system built of atoms. A Z-matrix is also known as an internal coordinate representation. It provides a description of each atom in a molecule in terms of its atomic number, bond length, bond angle, and dihedral angle, the so-called internal coordinates, although it is not always the case that a Z-matrix will give information regarding bonding since the matrix itself is based on a series of vectors describing atomic orientations in space. However, it is convenient to write a Z-matrix in terms of bond lengths, angles, and dihedrals since this will preserve the actual bonding characteristics. The name arises because the Z-matrix assigns the second atom along the Z axis from the first atom, which is at the origin. Z-matrices can be converted to Cartesian coordinates and back, as the structural information content is identical, the position and orientation in space, however is not meaning the Cartesian coordinates recovered will be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Density
In quantum chemistry, electron density or electronic density is the measure of the probability of an electron being present at an infinitesimal element of space surrounding any given point. It is a scalar quantity depending upon three spatial variables and is typically denoted as either \rho(\textbf r) or n(\textbf r). The density is determined, through definition, by the normalised N-electron wavefunction which itself depends upon 4N variables (3N spatial and N spin coordinates). Conversely, the density determines the wave function modulo up to a phase factor, providing the formal foundation of density functional theory. According to quantum mechanics, due to the uncertainty principle on an atomic scale the exact location of an electron cannot be predicted, only the probability of its being at a given position; therefore electrons in atoms and molecules act as if they are "smeared out" in space. For one-electron systems, the electron density at any point is proportional to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
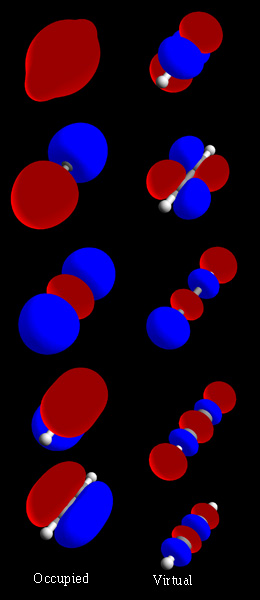
Molecular Orbitals
In chemistry, a molecular orbital is a mathematical function describing the location and wave-like behavior of an electron in a molecule. This function can be used to calculate chemical and physical properties such as the probability of finding an electron in any specific region. The terms ''atomic orbital'' and ''molecular orbital'' were introduced by Robert S. Mulliken in 1932 to mean ''one-electron orbital wave functions''. At an elementary level, they are used to describe the ''region'' of space in which a function has a significant amplitude. In an isolated atom, the orbital electrons' location is determined by functions called atomic orbitals. When multiple atoms combine chemically into a molecule, the electrons' locations are determined by the molecule as a whole, so the atomic orbitals combine to form molecular orbitals. The electrons from the constituent atoms occupy the molecular orbitals. Mathematically, molecular orbitals are an approximate solution to the Schrödin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PySCF
Python-based Simulations of Chemistry Framework (PySCF) is an ab initio computational chemistry program natively implemented in Python program language. The package aims to provide a simple, light-weight and efficient platform for quantum chemistry code developing and calculation. It provides various functions to do the Hartree–Fock, MP2, density functional theory, MCSCF, coupled cluster Coupled cluster (CC) is a numerical technique used for describing many-body systems. Its most common use is as one of several post-Hartree–Fock ab initio quantum chemistry methods in the field of computational chemistry, but it is also used in ... theory at non-relativistic level and 4-component relativistic Hartree–Fock theory. Although most functions are written in Python, the computation critical modules are intensively optimized in C. As a result, the package works as efficient as other C/ Fortran-based quantum chemistry program. PySCF is developed by Qiming Sun. PySCF2.0 is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linux
Linux ( or ) is a family of open-source Unix-like operating systems based on the Linux kernel, an operating system kernel first released on September 17, 1991, by Linus Torvalds. Linux is typically packaged as a Linux distribution, which includes the kernel and supporting system software and libraries, many of which are provided by the GNU Project. Many Linux distributions use the word "Linux" in their name, but the Free Software Foundation uses the name "GNU/Linux" to emphasize the importance of GNU software, causing some controversy. Popular Linux distributions include Debian, Fedora Linux, and Ubuntu, the latter of which itself consists of many different distributions and modifications, including Lubuntu and Xubuntu. Commercial distributions include Red Hat Enterprise Linux and SUSE Linux Enterprise. Desktop Linux distributions include a windowing system such as X11 or Wayland, and a desktop environment such as GNOME or KDE Plasma. Distributions intended for ser ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MOLPRO
MOLPRO is a software package used for accurate ''ab initio'' quantum chemistry calculations. It is developed by Peter Knowles at Cardiff University and Hans-Joachim Werner at Universität Stuttgart in collaboration with other authors. The emphasis in the program is on highly accurate computations, with extensive treatment of the electron correlation problem through the multireference configuration interaction, coupled cluster and associated methods. Integral-direct local electron correlation methods reduce the increase of the computational cost with molecular size. Accurate ab initio calculations can then be performed for larger molecules. With new explicitly correlated methods the basis set limit can be very closely approached. History Molpro was designed and maintained by Wilfried Meyer and Peter Pulay Peter Pulay (born September 20, 1941, in Veszprém, Hungary) is a theoretical chemist. He is the Roger B. Bost Distinguished Professor of Chemistry in the Department of Chemis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)
