|
Model Organism Database
Model organism databases (MODs) are biological databases, or knowledgebases, dedicated to the provision of in-depth biological data for intensively studied model organisms. MODs allow researchers to easily find background information on large sets of genes, plan experiments efficiently, combine their data with existing knowledge, and construct novel hypotheses. They allow users to analyse results and interpret datasets, and the data they generate are increasingly used to describe less well studied species. Where possible, MODs share common approaches to collect and represent biological information. For example, all MODs use the Gene Ontology (GO) to describe functions, processes and cellular locations of specific gene products. Projects also exist to enable software sharing for curation, visualization and querying between different MODs. Organismal diversity and varying user requirements however mean that MODs are often required to customize capture, display, and provision of data ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Databases
Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization (both cellular and chromosomal), clinical effects of mutations as well as similarities of biological sequences and structures. Biological databases can be classified by the kind of data they collect (see below). Broadly, there are molecular databases (for sequences, molecules, etc.), functional databases (for physiology, enzyme activities, phenotypes, ecology etc), taxonomic databases (for species and other taxonomic ranks), images and other media, or specimens (for museum collections etc.) Databases are important tools in assisting scientists to analyze and explain a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caenorhabditis Elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (rod-like) and Latin ''elegans'' (elegant). In 1900, Maupas initially named it '' Rhabditides elegans.'' Osche placed it in the subgenus ''Caenorhabditis'' in 1952, and in 1955, Dougherty raised ''Caenorhabditis'' to the status of genus. ''C. elegans'' is an unsegmented pseudocoelomate and lacks respiratory or circulatory systems. Most of these nematodes are hermaphrodites and a few are males. Males have specialised tails for mating that include spicules. In 1963, Sydney Brenner proposed research into ''C. elegans,'' primarily in the area of neuronal development. In 1974, he began research into the molecular and developmental biology of ''C. elegans'', which has since been extensively used as a model organism. It was the first mult ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Danio Rerio
The zebrafish (''Danio rerio'') is a freshwater fish belonging to the minnow family (Cyprinidae) of the order Cypriniformes. Native to South Asia, it is a popular aquarium fish, frequently sold under the trade name zebra danio (and thus often called a " tropical fish" although both tropical and subtropical). It is also found in private ponds. The zebrafish is an important and widely used vertebrate model organism in scientific research, for example in drug development, in particular pre-clinical development. It is also notable for its regenerative abilities, and has been modified by researchers to produce many transgenic strains. Taxonomy The zebrafish is a derived member of the genus ''Brachydanio'', of the family Cyprinidae. It has a sister-group relationship with ''Danio aesculapii''. Zebrafish are also closely related to the genus ''Devario'', as demonstrated by a phylogenetic tree of close species. Distribution Range The zebrafish is native to fresh water habit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SoyBase Database
SoyBase is a database created by the United States Department of Agriculture. It contains genetic information about soybeans. It includes genetic maps, information about Mendelian genetics and molecular data regarding genes and sequences. It was started in 1990 and is freely available to individuals and organizations worldwide. History SoyBase was instituted by the Corn Insects and Corn Genetics Research Unit (CICGRU) in Ames, Iowa as a central repository for the soybean genetics community's published information. Originally, the database concentrated on genetic information such as genetic linkage maps and other Mendalian information. SoyBase genetic maps are a manually-curated composite of all published mapping and QTL studies, and thus provide a species level view of markers and QTL. In 2010 the soybean genome sequence was released along with gene models and many other types of genome annotations that were integrated in to SoyBase. SoyBase genetic linkage maps were integral ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine Soja
''Glycine soja'', or wild soybean (previously ''G. ussuriensis'') is an annual plant in the legume family. It is the closest living relative of soybean The soybean, soy bean, or soya bean (''Glycine max'') is a species of legume native to East Asia, widely grown for its edible bean, which has numerous uses. Traditional unfermented food uses of soybeans include soy milk, from which tofu ..., an important crop. The plant is native to eastern China, Japan, Korea and far-eastern Russia. References External links *Plants for a Future soja Crops originating from China {{Phaseoleae-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Maize
Maize ( ; ''Zea mays'' subsp. ''mays'', from es, maíz after tnq, mahiz), also known as corn (North American and Australian English), is a cereal grain first domesticated by indigenous peoples in southern Mexico about 10,000 years ago. The leafy stalk of the plant produces pollen inflorescences (or "tassels") and separate ovuliferous inflorescences called ears that when fertilized yield kernels or seeds, which are fruits. The term ''maize'' is preferred in formal, scientific, and international usage as a common name because it refers specifically to this one grain, unlike ''corn'', which has a complex variety of meanings that vary by context and geographic region. Maize has become a staple food in many parts of the world, with the total production of maize surpassing that of wheat or rice. In addition to being consumed directly by humans (often in the form of masa), maize is also used for corn ethanol, animal feed and other maize products, such as corn starch and corn sy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The Arabidopsis Information Resource
The Arabidopsis Information Resource (TAIR) is a community resource and online model organism database of genetic and molecular biology data for the model plant ''Arabidopsis thaliana ''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small flowering plant native to Eurasia and Africa. ''A. thaliana'' is considered a weed; it is found along the shoulders of roads and in disturbed land. A winter ...'', commonly known as mouse-ear cress. TAIR integrates information about the Arabidopsis genome, genes, gene products, natural variants, mutant alleles and plant phenotypes and research literature. Data in TAIR can be retrieved using simple and advanced searches, bulk query and download tools, and in collections of prepared text files. The Arabidopsis genome and annotations can be visualized using the interactive SeqViewer and GBrowse tools. TAIR’s biocurators are responsible for acquiring and integrating data from the research literature (functional ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arabidopsis Thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small flowering plant native to Eurasia and Africa. ''A. thaliana'' is considered a weed; it is found along the shoulders of roads and in disturbed land. A winter annual with a relatively short lifecycle, ''A. thaliana'' is a popular model organism in plant biology and genetics. For a complex multicellular eukaryote, ''A. thaliana'' has a relatively small genome around 135 megabase pairs. It was the first plant to have its genome sequenced, and is a popular tool for understanding the molecular biology of many plant traits, including flower development and light sensing. Description ''Arabidopsis thaliana'' is an annual (rarely biennial) plant, usually growing to 20–25 cm tall. The leaves form a rosette at the base of the plant, with a few leaves also on the flowering stem. The basal leaves are green to slightly purplish in color, 1.5–5 cm long, and 2–10 mm broad, with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
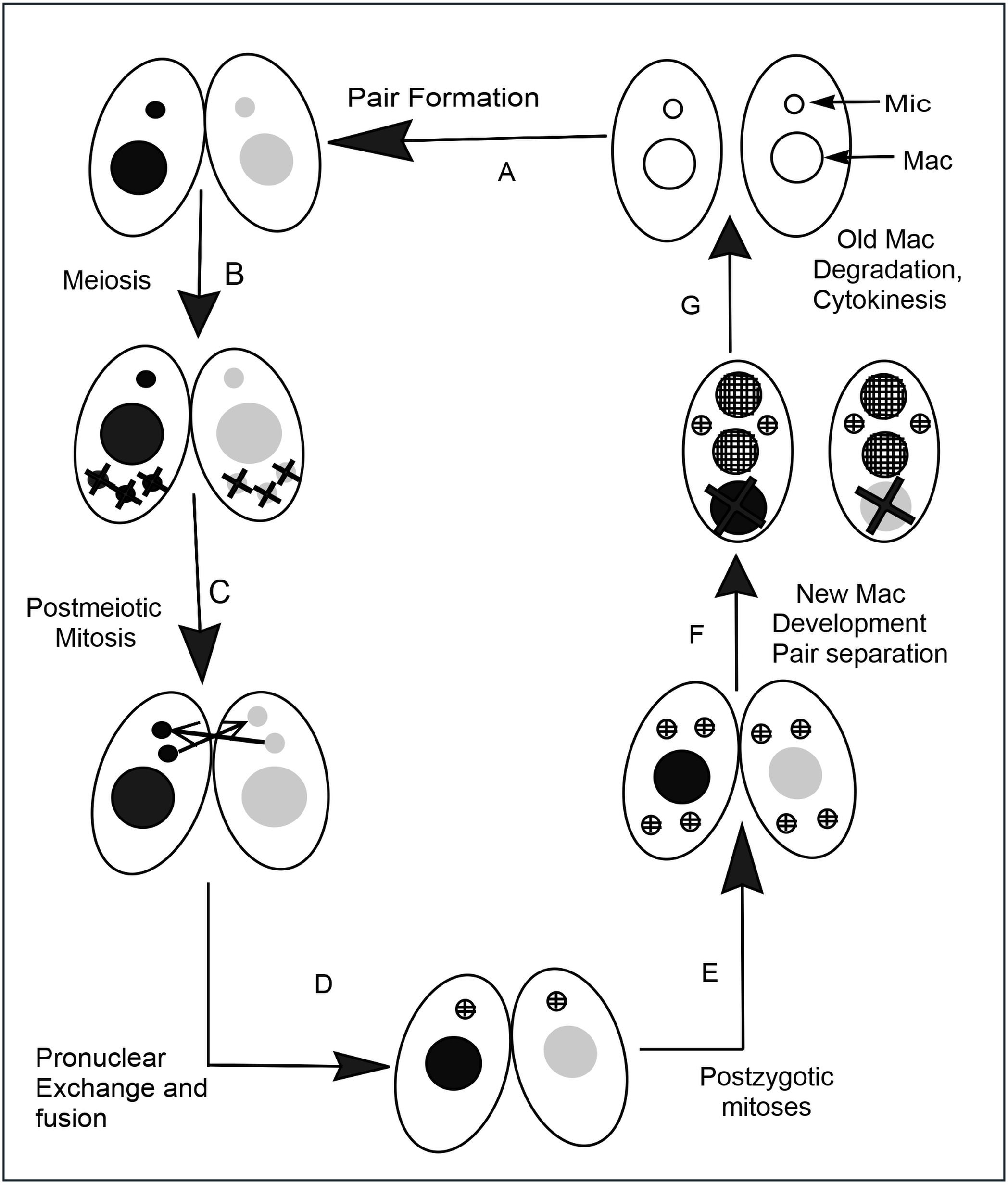
Tetrahymena Genome Database
''Tetrahymena'', a unicellular eukaryote, is a genus of free-living ciliates. The genus Tetrahymena is the most widely studied member of its phylum. It can produce, store and react with different types of hormones. Tetrahymena cells can recognize both related and hostile cells. They can also switch from commensalistic to pathogenic modes of survival. They are common in freshwater lakes, ponds, and streams. ''Tetrahymena'' species used as model organisms in biomedical research are ''T. thermophila'' and '' T. pyriformis''. ''T. thermophila'': a model organism in experimental biology As a ciliated protozoan, ''Tetrahymena thermophila'' exhibits nuclear dimorphism: two types of cell nuclei. They have a bigger, non-germline macronucleus and a small, germline micronucleus in each cell at the same time and these two carry out different functions with distinct cytological and biological properties. This unique versatility allows scientists to use ''Tetrahymena'' to ide ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrahymena Thermophila
''Tetrahymena thermophila'' is a species of Ciliophora in the family Tetrahymenidae. It is a free living protozoa and occurs in fresh water. There is little information on the ecology and natural history of this species, but it is the most widely known and widely studied species in the genus ''Tetrahymena''. The species has been used as a model organism for molecular and cellular biology. It has also helped in the discovery of new genes as well as help to understand the mechanisms of certain genes functions. Studies on this species have contributed to major discoveries in biology. For example, the MAT locus found in this species has provided a foundation for the evolution of mating systems. The species was first considered to be ''Tetrahymena pyriformis''. '' T. malaccensis'' is the closest relative to''T. thermophila''. Characteristics It is about 50 μm long. One famous trait this species is known for is that has 7 different mating types, unlike most eukaryotic organi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DictyBase
dictyBase is an online bioinformatics database for the model organism ''Dictyostelium discoideum ''Dictyostelium discoideum'' is a species of soil-dwelling amoeba belonging to the phylum Amoebozoa, infraphylum Mycetozoa. Commonly referred to as slime mold, ''D. discoideum'' is a eukaryote that transitions from a collection of unicellular ...''. Tools dictyBase offers many ways of searching and retrieving data from the database: * dictyMart - a tool for retrieving varied information on many genes (or the sequences of those genes). * Genome Browser - browse the genes of ''D. discoideum'' in their genomic context. References External links dictyBase Developmental biology Model organism databases Mycetozoa {{Database-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dictyostelium Discoideum
''Dictyostelium discoideum'' is a species of soil-dwelling amoeba belonging to the phylum Amoebozoa, infraphylum Mycetozoa. Commonly referred to as slime mold, ''D. discoideum'' is a eukaryote that transitions from a collection of unicellular amoebae into a multicellular slug and then into a fruiting body within its lifetime. Its unique asexual lifecycle consists of four stages: vegetative, aggregation, migration, and culmination. The lifecycle of ''D. discoideum'' is relatively short, which allows for timely viewing of all stages. The cells involved in the lifecycle undergo movement, chemical signaling, and development, which are applicable to human cancer research. The simplicity of its lifecycle makes ''D. discoideum'' a valuable model organism to study genetic, cellular, and biochemical processes in other organisms. Natural habitat and diet In the wild, ''D. discoideum'' can be found in soil and moist leaf litter. Its primary diet consists of bacteria, such as ''Escherichia ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |