|
MG-RAST
MG-RAST is an open-source web application server that suggests automatic phylogenetic and functional analysis of metagenomes. It is also one of the biggest repositories for metagenomic data. The name is an abbreviation of ''Metagenomic Rapid Annotations using Subsystems Technology''. The pipeline automatically produces functional assignments to the sequences that belong to the metagenome by performing sequence comparisons to databases in both nucleotide and amino-acid levels. The applications supply phylogenetic and functional assignments of the metagenome being analysed, as well as tools for comparing different metagenomes. It also provides RESTful APIfor programmatic access. The server was created and maintained by Argonne National Laboratory from the University of Chicago. In December 29 of 2016, the system had analyzed 60 terabase-pairs of data from more than 150,000 data sets. Among the analyzed data sets, more than 23,000 are available to the public. Currently, the computa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenome
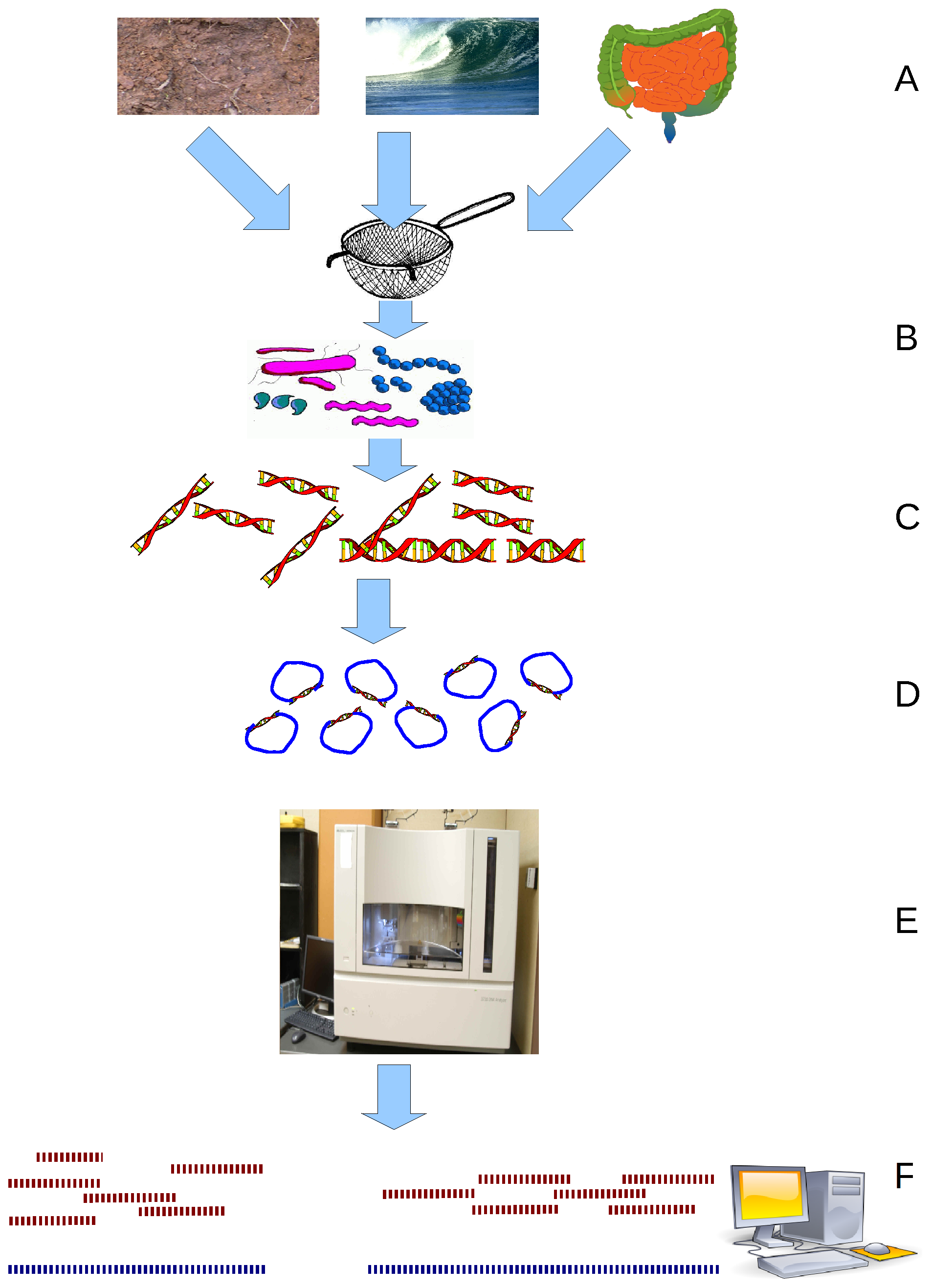
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Argonne National Laboratory
Argonne National Laboratory is a science and engineering research United States Department of Energy National Labs, national laboratory operated by University of Chicago, UChicago Argonne LLC for the United States Department of Energy. The facility is located in Lemont, Illinois, outside of Chicago, and is the largest national laboratory by size and scope in the Midwest. Argonne had its beginnings in the Metallurgical Laboratory of the University of Chicago, formed in part to carry out Enrico Fermi's work on nuclear reactors for the Manhattan Project during World War II. After the war, it was designated as the first national laboratory in the United States on July 1, 1946. In the post-war era the lab focused primarily on non-weapon related nuclear physics, designing and building the first power-producing nuclear reactors, helping design the reactors used by the United States' nuclear navy, and a wide variety of similar projects. In 1994, the lab's nuclear mission ended, and today ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by Ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
OpenMP
OpenMP (Open Multi-Processing) is an application programming interface (API) that supports multi-platform shared-memory multiprocessing programming in C, C++, and Fortran, on many platforms, instruction-set architectures and operating systems, including Solaris, AIX, FreeBSD, HP-UX, Linux, macOS, and Windows. It consists of a set of compiler directives, library routines, and environment variables that influence run-time behavior. OpenMP is managed by the nonprofit technology consortium ''OpenMP Architecture Review Board'' (or ''OpenMP ARB''), jointly defined by a broad swath of leading computer hardware and software vendors, including Arm, AMD, IBM, Intel, Cray, HP, Fujitsu, Nvidia, NEC, Red Hat, Texas Instruments, and Oracle Corporation. OpenMP uses a portable, scalable model that gives programmers a simple and flexible interface for developing parallel applications for platforms ranging from the standard desktop computer to the supercomputer. An application built wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
QIIME
QIIME (an abbreviation for ''Quantitative Insights Into Microbial Ecology'') is a bioinformatic pipeline designated for the task of analysing microbial communities that were sampled through marker gene (e.g. 16S or 18S rRNA genes) amplicon sequencing. In its heart the pipeline performs quality control over the input sequencing reads, clusters the marker gene nucleotide sequences at a requested phylogenetic level (e.g. 97% for species level) into OTUs (operational taxonomic units) or sequence variants and taxonomically annotates them by looking for similar sequences in a reference taxonomic database. The main output from the QIIME pipeline is a feature table, which describes the abundances of each OTU or sequence variant in each sample. Additional tools that are relevant to ecological aspects of the samples being investigated are also provided within the pipeline, including rarefaction, alpha diversity and beta diversity calculations, visualizations such as principle coordinates ana ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UCLUST
UCLUST is an algorithm designed to cluster nucleotide or amino-acid sequences into clusters based on sequence similarity. The algorithm was published in 2010 and implemented in a program also named UCLUST. The algorithm is described by the author as following two simple clustering criteria, in regard to the requested similarity threshold T. The first criterion states that any given cluster's centroid sequence will have a similarity smaller than T to any other clusters' centroid sequence. The second criterion states that each member sequence in a given cluster will have similarity to the cluster's centroid sequence that is equal or greater than T. UCLUST algorithm is a greedy one. As a result, the order of the sequences in the input file will affect the resulting clusters and their quality. For this reason, it is advised that the sequences will be sorted before entering clustering stage. The program UCLUST is equipped with some options to sort the input sequences prior to clustering ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SILVA Ribosomal RNA Database
SILVA is a ribosomal RNA database established in collaboration between the Microbial Genomics Group at the Max Planck Institute for Marine Microbiology in Bremen, Germany, the Department of Microbiology at the Technical University Munich, and Ribocon. Release 117 of the database (January 2014) held more than 4,000,000 small subunit (SSU - 16S/18S) and 400,000 large subunit In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of ty ... (LSU - 23S/28S) sequences. Sequences are provided as files for the ARB software environment. See also * List of biological databases#RNA databases References {{reflist, refs= Marc P. Hoeppner, Lars E. Barquist, Paul P. Gardner (2014)An Introduction to RNA Databases ''Methods in Molecular Biology'' 1097: 107–123. {{doi, 10.1007/978-1-62703-709-9_6. [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAT (bioinformatics)
BLAT (BLAST-like alignment tool) is a pairwise sequence alignment algorithm that was developed by Jim Kent at the University of California Santa Cruz (UCSC) in the early 2000s to assist in the assembly and annotation of the human genome. It was designed primarily to decrease the time needed to align millions of mouse genomic reads and expressed sequence tags against the human genome sequence. The alignment tools of the time were not capable of performing these operations in a manner that would allow a regular update of the human genome assembly. Compared to pre-existing tools, BLAT was ~500 times faster with performing mRNA/ DNA alignments and ~50 times faster with protein/protein alignments. Overview BLAT is one of multiple algorithms developed for the analysis and comparison of biological sequences such as DNA, RNA and proteins, with a primary goal of inferring homology in order to discover biological function of genomic sequences. It is not guaranteed to find the mathematicall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Prediction
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functional elements such as regulatory regions. Gene finding is one of the first and most important steps in understanding the genome of a species once it has been sequenced. In its earliest days, "gene finding" was based on painstaking experimentation on living cells and organisms. Statistical analysis of the rates of homologous recombination of several different genes could determine their order on a certain chromosome, and information from many such experiments could be combined to create a genetic map specifying the rough location of known genes relative to each other. Today, with comprehensive genome sequence and powerful computational resources at the disposal of the research community, gene finding has been redefined as a largely computatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |