|
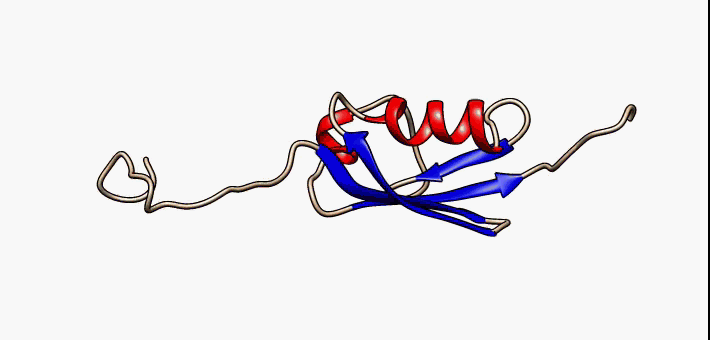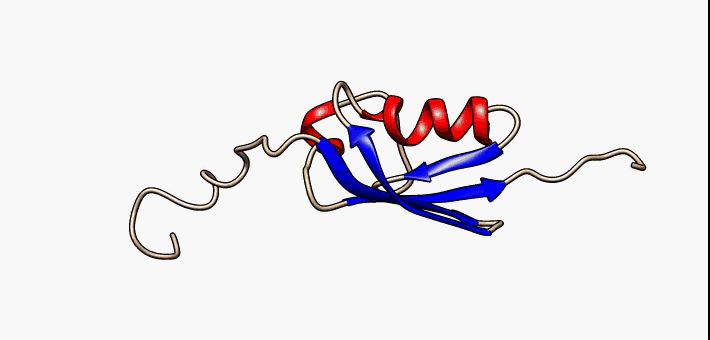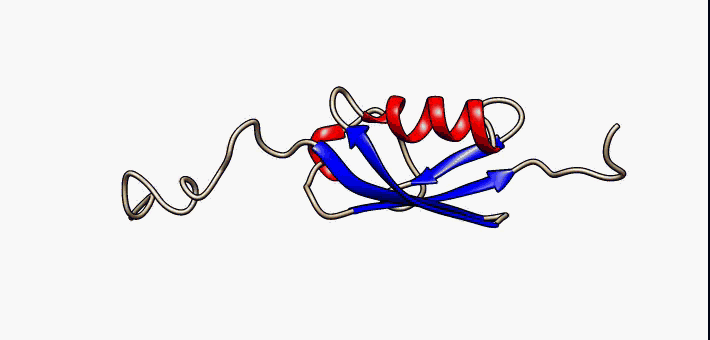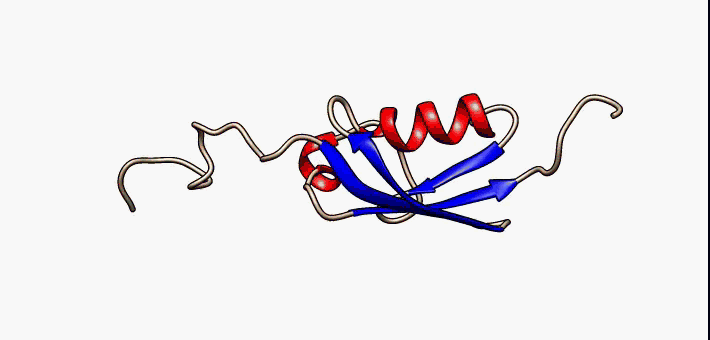
Molecular Recognition Feature
Molecular recognition features (MoRFs) are small (10-70 residues) intrinsically disordered regions in proteins that undergo a disorder-to-order transition upon binding to their partners. MoRFs are implicated in protein-protein interactions, which serve as the initial step in molecular recognition. MoRFs are disordered prior to binding to their partners, whereas they form a common 3D structure after interacting with their partners. As MoRF regions tend to resemble disordered proteins with some characteristics of ordered proteins, they can be classified as existing in an extended semi-disordered state. Categorization MoRFs can be separated in 4 categories according to the shape they form once bound to their partners. The categories are: * α-MoRFs (when they form alpha-helixes) * β-MoRFs (when they form beta-sheets) * irregular-MoRFs (when they don't form any shape) * complex-MoRFs (combination of the above categories) MoRF predictors Determining protein structures experime ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Proteins
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent interactions that are highly cooperative and dynamic, lending them importance in DNA regulation and in cell signaling. Many IDPs can also adopt a fixed three-dimensional structure after bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Recognition
The term molecular recognition refers to the specific interaction between two or more molecules through noncovalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, π-π interactions, halogen bonding, or resonant interaction effects. In addition to these ''direct'' interactions, solvents can play a dominant ''indirect'' role in driving molecular recognition in solution. The host and guest involved in molecular recognition exhibit molecular complementarity. Exceptions are molecular containers, including e.g. nanotubes, in which portals essentially control selectivity. Biological systems Molecular recognition plays an important role in biological systems and is observed in between receptor-ligand, antigen-antibody, DNA-protein, sugar- lectin, RNA-ribosome, etc. An important example of molecular recognition is the antibiotic vancomycin that selectively binds with the peptides with terminal D-alanyl-D-alanine in bacterial cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary structures ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
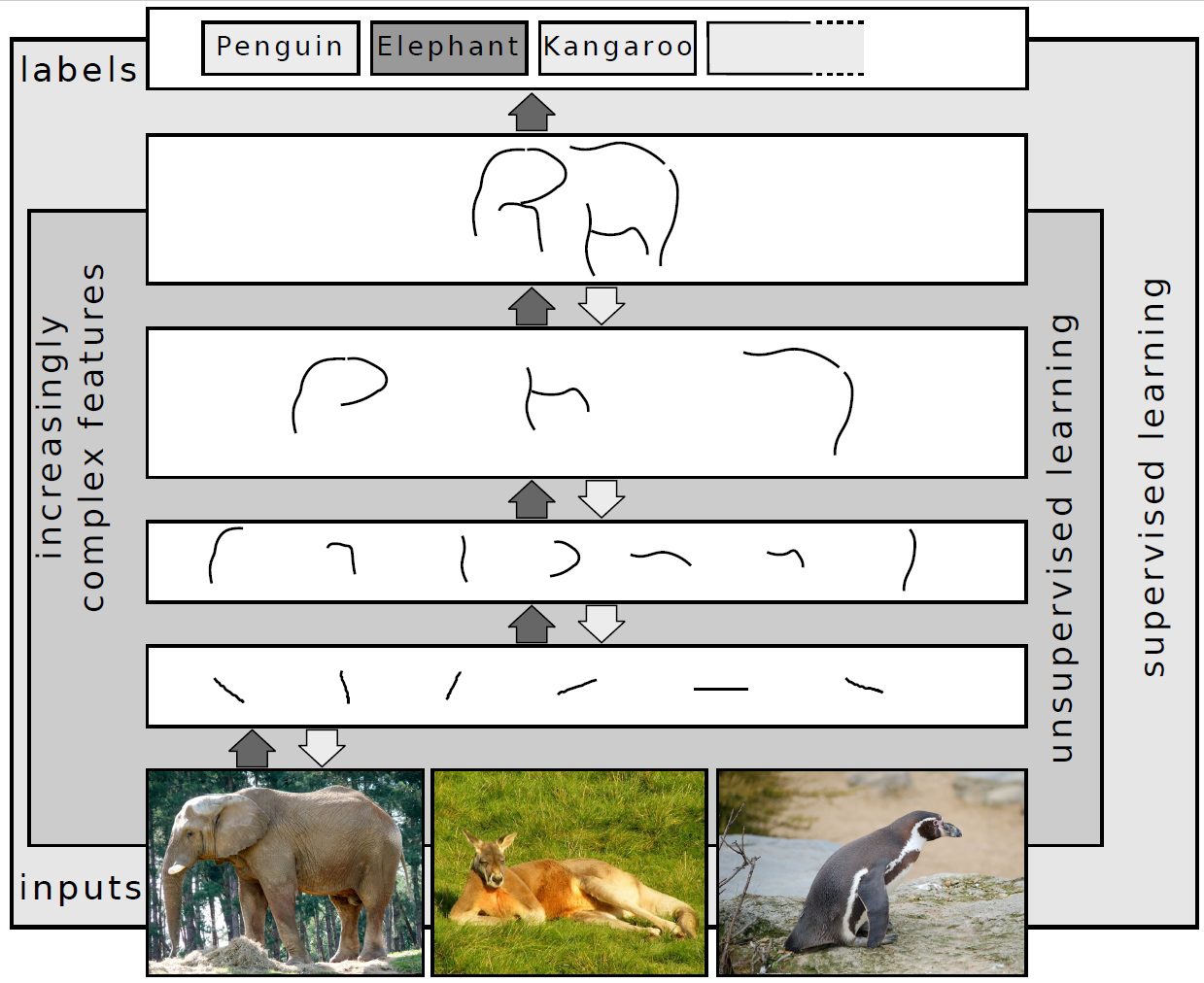
Deep Learning
Deep learning (also known as deep structured learning) is part of a broader family of machine learning methods based on artificial neural networks with representation learning. Learning can be supervised, semi-supervised or unsupervised. Deep-learning architectures such as deep neural networks, deep belief networks, deep reinforcement learning, recurrent neural networks, convolutional neural networks and Transformers have been applied to fields including computer vision, speech recognition, natural language processing, machine translation, bioinformatics, drug design, medical image analysis, Climatology, climate science, material inspection and board game programs, where they have produced results comparable to and in some cases surpassing human expert performance. Artificial neural networks (ANNs) were inspired by information processing and distributed communication nodes in biological systems. ANNs have various differences from biological brains. Specifically, artificial ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |