|
Microarray Chip
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The " gene chip" industry started to grow significantly after the 1995 '' Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophisticat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Venn Diagram For BioMEMS, LOC, And MTAS
Venn is a surname and a given name. It may refer to: Given name * Venn Eyre (died 1777), Archdeacon of Carlisle, Cumbria, England * Venn Pilcher (1879–1961), Anglican bishop, writer, and translator of hymns * Venn Young (1929–1993), New Zealand politician Surname * Albert Venn (1867–1908), American lacrosse player * Anne Venn (1620s–1654), English religious radical and diarist * Blair Venn, Australian actor * Charles Venn (born 1973), British actor * Harry Venn (1844–1908), Australian politician * Henry Venn (Church Missionary Society) the younger (1796-1873), secretary of the Church Missionary Society, grandson of Henry Venn * Henry Venn (Clapham Sect) the elder (1725–1797), English evangelical minister * Horace Venn (1892–1953), English cricketer * John Venn (1834–1923), British logician and the inventor of Venn diagrams, son of Henry Venn the younger * John Venn (academic) (died 1687), English academic administrator * John Venn (politician) (1586–1650), Engli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was inv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
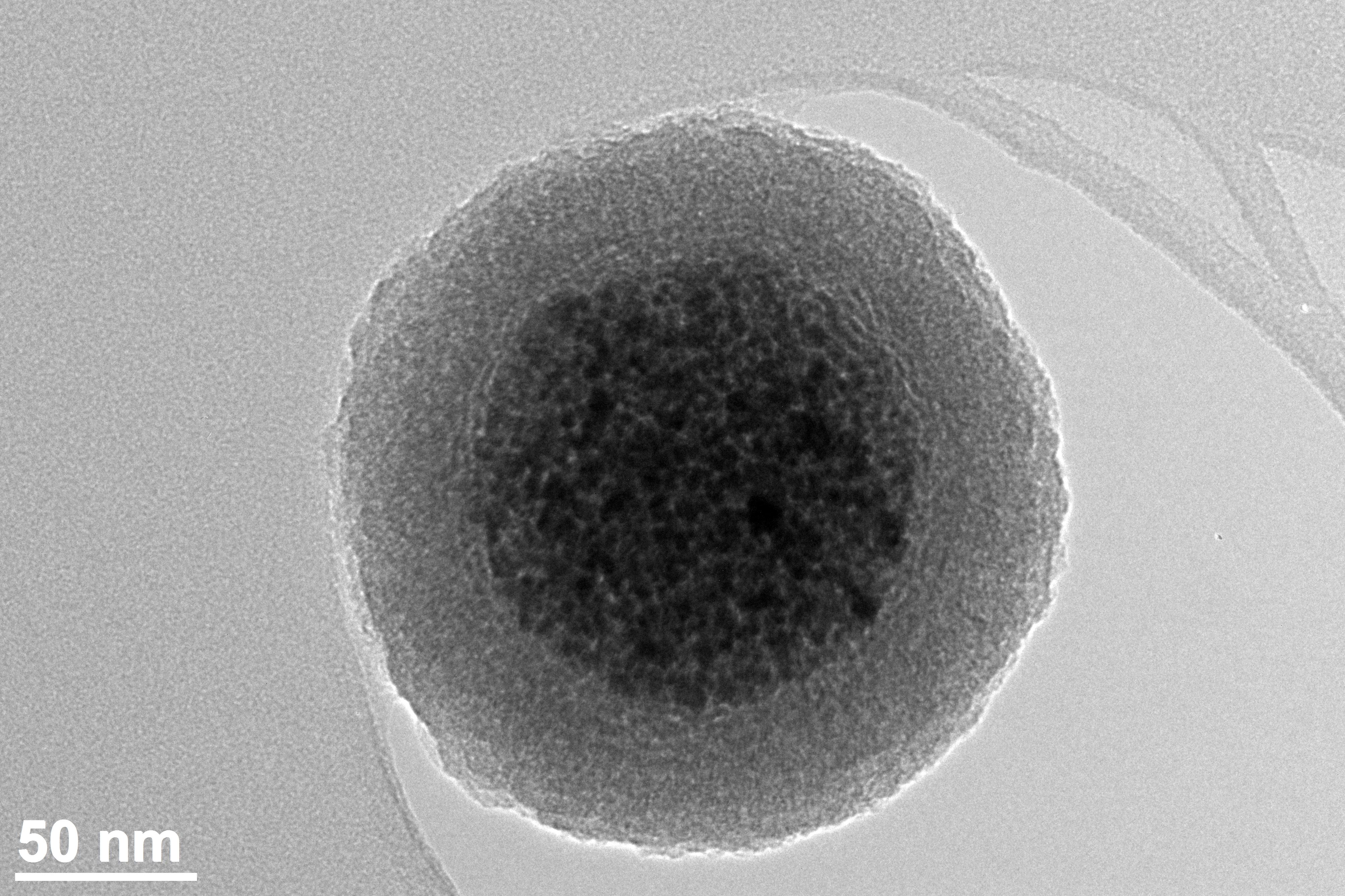
Magnetic Nanoparticles
Magnetic nanoparticles are a class of nanoparticle that can be manipulated using magnetic fields. Such particles commonly consist of two components, a magnetic material, often iron, nickel and cobalt, and a chemical component that has functionality. While nanoparticles are smaller than 1 micrometer in diameter (typically 1–100 nanometers), the larger microbeads are 0.5–500 micrometer in diameter. Magnetic nanoparticle clusters that are composed of a number of individual magnetic nanoparticles are known as magnetic nanobeads with a diameter of 50–200 nanometers. Magnetic nanoparticle clusters are a basis for their further magnetic assembly into magnetic nanochains. The magnetic nanoparticles have been the focus of much research recently because they possess attractive properties which could see potential use in catalysis including nanomaterial-based catalysts, biomedicine and tissue specific targeting, antimicrobial agents, magnetically tunable colloidal photonic crystals, mic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Phase Protein Lysate Microarray
A reverse phase protein lysate microarray (RPMA) is a protein microarray designed as a dot-blot platform that allows measurement of protein expression levels in a large number of biological samples simultaneously in a quantitative manner when high-quality antibodies are available. Technically, minuscule amounts of (a) cellular lysates, from intact cells or laser capture microdissected cells, (b) body fluids such as serum, CSF, urine, vitreous, saliva, etc., are immobilized on individual spots on a microarray that is then incubated with a single specific antibody to detect expression of the target protein across many samples. A summary video of RPPA is available. One microarray, depending on the design, can accommodate hundreds to thousands of samples that are printed in a series of replicates. Detection is performed using either a primary or a secondary labeled antibody by chemiluminescent, fluorescent or colorimetric assays. The array is then imaged and the obtained data is quant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype Microarray
The phenotype microarray approach is a technology for high-throughput phenotyping of cells. A phenotype microarray system enables one to monitor simultaneously the phenotypic reaction of cells to environmental challenges or exogenous compounds in a high-throughput manner. The phenotypic reactions are recorded as either end-point measurements or respiration kinetics similar to growth curves. Usages High-throughput phenotypic testing is increasingly important for exploring the biology of bacteria, fungi, yeasts, and animal cell lines such as human cancer cells. Just as DNA microarrays and proteomic technologies have made it possible to assay the expression level of thousands of genes or proteins all a once, phenotype microarrays (PMs) make it possible to quantitatively measure thousands of cellular phenotypes simultaneously. The approach also offers potential for testing gene function and improving genome annotation. In contrast to many of the hitherto available molecular high- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycan Array
Glycan arrays, like that offered by the Consortium for Functional Glycomics (CFG), National Center for Functional Glycomics (NCFG) anZ Biotech, LLC contain carbohydrate compounds that can be screened with lectins, antibodies or cell receptors to define carbohydrate specificity and identify ligands. Glycan array screening works in much the same way as other microarray that is used for instance to study gene expression DNA microarrays or protein interaction Protein microarrays. Glycan arrays are composed of various oligosaccharides and/or polysaccharides immobilised on a solid support in a spatially-defined arrangement. This technology provides the means of studying glycan-protein interactions in a high-throughput environment. These natural or synthetic (see carbohydrate synthesis) glycans are then incubated with any glycan-binding protein such as lectins, cell surface receptors or possibly a whole organism such as a virus. Binding is quantified using fluorescence-based detection met ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody Microarray
An antibody microarray (also known as antibody array) is a specific form of protein microarray. In this technology, a collection of captured antibodies are spotted and fixed on a solid surface such as glass, plastic, membrane, or silicon chip, and the interaction between the antibody and its target antigen is detected. Antibody microarrays are often used for detecting protein expression from various biofluids including serum, plasma and cell or tissue lysates. Antibody arrays may be used for both basic research and medical and diagnostic applications. Background The concept and methodology of antibody microarrays were first introduced by Tse Wen Chang in 1983 in a scientific publication and a series of patents, when he was working at Centocor in Malvern, Pennsylvania. Chang coined the term “antibody matrix” and discussed “array” arrangement of minute antibody spots on small glass or plastic surfaces. He demonstrated that a 10×10 (100 in total) and 20×20 (400 in total) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Compound Microarray
A chemical compound microarray is a collection of organic chemical compounds spotted on a solid surface, such as glass and plastic. This microarray format is very similar to DNA microarray, protein microarray and antibody microarray. In chemical genetics research, they are routinely used for searching proteins that bind with specific chemical compounds, and in general drug discovery research, they provide a multiplex way to search potential drugs for therapeutic targets. There are three different forms of chemical compound microarrays based on the fabrication method. The first form is to covalently immobilize the organic compounds on the solid surface with diverse linking techniques; this platform is usually called Small Molecule Microarray, which is invented and advanced by Dr. Stuart Schreiber and colleague The second form is to spot and dry organic compounds on the solid surface without immobilization, this platform has a commercial name as Micro Arrayed Compound Screening (μAR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Microarray
A cellular microarray (or cell microarray) is a laboratory tool that allows for the multiplex interrogation of living cells on the surface of a solid support. The support, sometimes called a "chip", is spotted with varying materials, such as antibodies, proteins, or lipids, which can interact with the cells, leading to their capture on specific spots. Combinations of different materials can be spotted in a given area, allowing not only cellular capture, when a specific interaction exists, but also the triggering of a cellular response, change in phenotype, or detection of a response from the cell, such as a specific secreted factor. There are a large number of types of cellular microarrays: # Reverse transfection cell microarrays. David M. Sabatini's laboratory developed reverse-transfection cell microarrays at the Whitehead Institute, publishing their work in 2001. # PMHC Cellular Microarrays PMHC cellular microarrays are a type of cellular microarray that has been spotted with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tissue Microarray
Tissue microarrays (also TMAs) consist of paraffin blocks in which up to 1000 separate tissue cores are assembled in array fashion to allow multiplex histological analysis. History The major limitations in molecular clinical analysis of tissues include the cumbersome nature of procedures, limited availability of diagnostic reagents and limited patient sample size. The technique of tissue microarray was developed to address these issues. Multi-tissue blocks were first introduced by H. Battifora in 1986 with his so-called “multitumor (sausage) tissue block" and modified in 1990 with its improvement, "the checkerboard tissue block" . In 1998, J. Kononen and collaborators developed the current technique, which uses a novel sampling approach to produce tissues of regular size and shape that can be more densely and precisely arrayed. Procedure In the tissue microarray technique, a hollow needle is used to remove tissue cores as small as 0.6 mm in diameter from regions of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |