|
Mfsd14b
Major facilitator superfamily domain containing 14B (MFSD14B, HIATL1) is an atypical solute carrier of MFS type. It locates to intracellular membranes. HGNC The HUGO Gene Nomenclature Committee (HGNC) is a committee of the Human Genome Organisation (HUGO) that sets the standards for human gene nomenclature. The HGNC approves a ''unique'' and ''meaningful'' name for every known human gene, based on a q ...:23376 TCDB: 2.A.1.2.30 MFSD14B cluster to AMTF1, together with MFSD9, MFSD10 and MFSD14A. References {{Reflist Protein superfamilies Solute carrier family Transmembrane proteins Transport proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Major Facilitator Superfamily
The major facilitator superfamily (MFS) is a superfamily of membrane transport proteins that facilitate movement of small solutes across cell membranes in response to chemiosmotic gradients. Function The major facilitator superfamily (MFS) are membrane proteins which are expressed ubiquitously in all kingdoms of life for the import or export of target substrates. The MFS family was originally believed to function primarily in the uptake of sugars but subsequent studies revealed that drugs, metabolites, oligosaccharides, amino acids and oxyanions were all transported by MFS family members. These protein energetically drive transport utilizing the electrochemical gradient of the target substrate (uniporter), or act as a cotransporter where transport is coupled to the movement of a second substrate. Fold The basic fold of the MFS transporter is built around 12, or in some cases, 14 transmembrane helices (TMH), with two 6- (or 7- ) helix bundles formed by the N and C terminal homo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Atypical MFS Transporter Family
Atypical Solute Carrier Families (Atypical SLCs) are novel plausible secondary active or facilitative transporter proteins that share ancestral background with the known solute carrier families (SLCs). However, they have not been assigned a name according to the SLC root system, or been classified into any of the existing SLC families. Atypical major facilitator superfamily transport families Most atypical SLCs are families within the major facilitator superfamily (MFS). These atypical SLCs are plausible secondary active or facilitative transporter proteins that share ancestry with the known solute carriers. They are, however, not named according to the SLC root system, or classified into any of the existing SLC families. ATMFs are categorised based on their sequence similarity and phylogenetic closeness. Some Atypical SLC of MFS type are: OCA2, CLN3, SPNS1, SPNS2, SPNS3, SV2A, SV2B, SV2C, SVOP, SVOPL, MFSD1, MFSD2A, MFSD2B, MFSD3, MFSD4A, MFSD4B, MFSD5, MFSD6, MFSD6L, M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Major Facilitator Superfamily Domain Containing 14a
Major facilitator superfamily domain containing 14A (MFSD14A, HIAT1) is a protein that in humans is encoded by the MFSD14A gene. MFSD14A is an atypical solute carrier of MFS type. HGNC:23363 MFSD14A cluster to AMTF1, together with MFSD9, MFSD10 and MFSD14B Major facilitator superfamily domain containing 14B (MFSD14B, HIATL1) is an atypical solute carrier of MFS type. It locates to intracellular membranes. HGNC The HUGO Gene Nomenclature Committee (HGNC) is a committee of the Human Genome Organisa .... References {{gene-1-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HUGO Gene Nomenclature Committee
The HUGO Gene Nomenclature Committee (HGNC) is a committee of the Human Genome Organisation (HUGO) that sets the standards for human gene nomenclature. The HGNC approves a ''unique'' and ''meaningful'' name for every known human gene, based on a query of experts. In addition to the name, which is usually 1 to 10 words long, the HGNC also assigns a symbol (a short group of characters) to every gene. As with an SI symbol, a gene symbol is like an abbreviation but is more than that, being a second unique name that can stand on its own just as much as substitute for the longer name. It may not necessarily "stand for" the initials of the name, although many gene symbols do reflect that origin. Purpose Especially gene abbreviations/symbols but also full gene names are often not specific for a single gene. A marked example is CAP which can refer to any of 6 different genes (BRD4'', CAP1'', HACD1'', LNPEP'', SERPINB6'', and SORBS1''). The HGNC short gene names, or gene symbols, unlike ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Superfamilies
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred (see homology). Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent (due to low sequence similarity). Superfamilies typically contain several protein families which show sequence similarity within each family. The term ''protein clan'' is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems. Identification Superfamilies of proteins are identified using a number of methods. Closely related members can be identified by different methods to those needed to group the most evolutionarily divergent members. Sequence similarity Historically, the similarity of different amino acid sequences has been the most common method of inferring homology. Sequence similarity is consid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
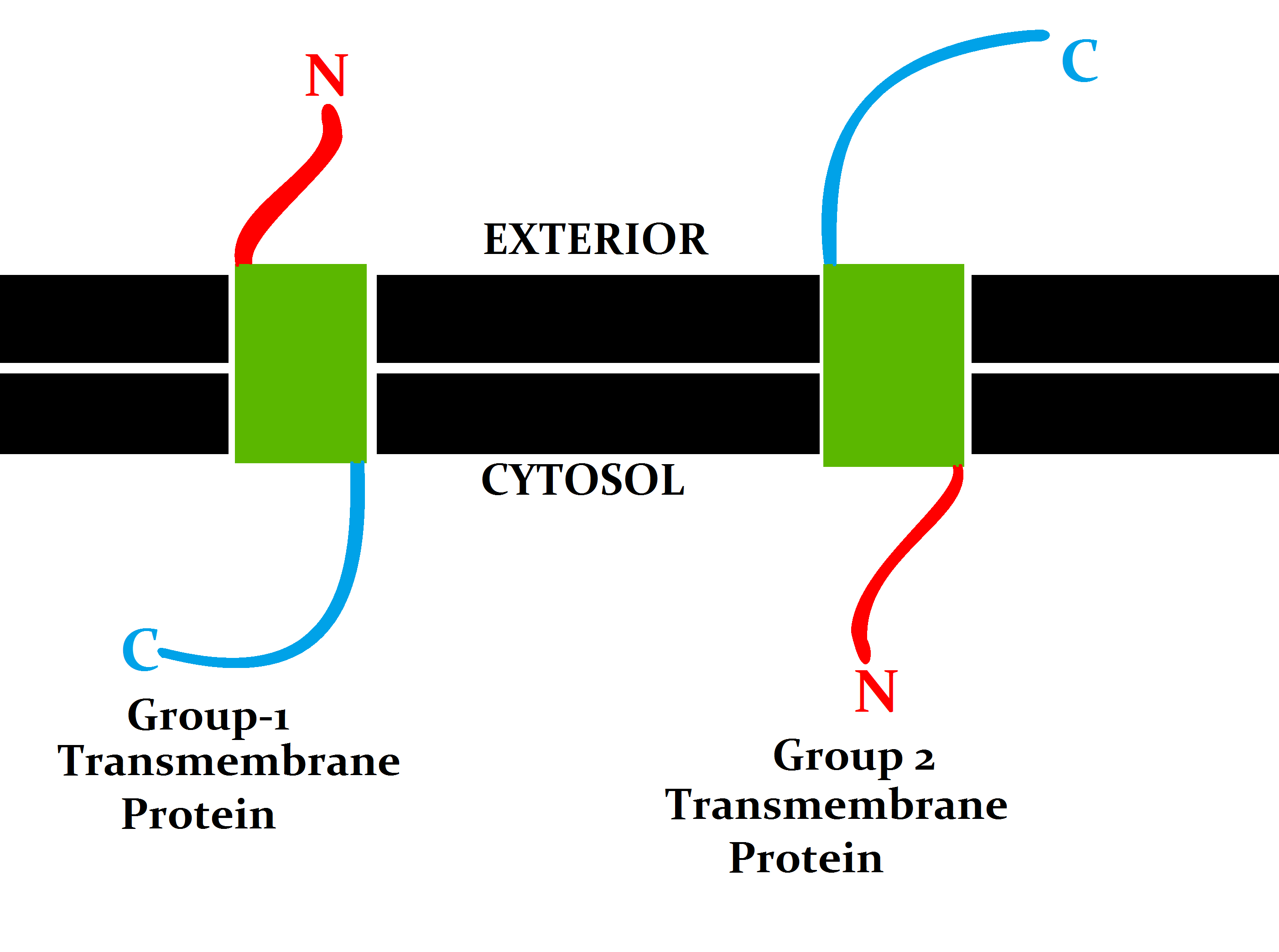
Transmembrane Proteins
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |