|
Linkage Disequilibrium Score Regression
In statistical genetics, linkage disequilibrium score regression (LDSR or LDSC) is a technique that aims to quantify the separate contributions of polygenic effects and various confounding factors, such as population stratification, based on summary statistics from genome-wide association studies (GWASs). The approach involves using regression analysis to examine the relationship between linkage disequilibrium scores and the test statistics of the single-nucleotide polymorphisms (SNPs) from the GWAS. Here, the "linkage disequilibrium score" for a SNP "is the sum of LD ''r2'' measured with all other SNPs". LDSC can be used to produce SNP-based heritability estimates, to partition this heritability into separate categories, and to calculate genetic correlations between separate phenotypes. Because the LDSC approach relies only on summary statistics from an entire GWAS, it can be used efficiently even with very large sample sizes. In LDSC, genetic correlations are calculated based on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Statistical Genetics
Statistical genetics is a scientific field concerned with the development and application of statistical methods for drawing inferences from genetic data. The term is most commonly used in the context of human genetics. Research in statistical genetics generally involves developing theory or methodology to support research in one of three related areas: *population genetics - Study of evolutionary processes affecting genetic variation between organisms *genetic epidemiology - Studying effects of genes on diseases *quantitative genetics - Studying the effects of genes on 'normal' phenotypes Statistical geneticists tend to collaborate closely with geneticists, molecular biologists, clinicians and bioinformaticians. Statistical genetics is a type of computational biology Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heritability
Heritability is a statistic used in the fields of breeding and genetics that estimates the degree of ''variation'' in a phenotypic trait in a population that is due to genetic variation between individuals in that population. The concept of heritability can be expressed in the form of the following question: "What is the proportion of the variation in a given trait within a population that is ''not'' explained by the environment or random chance?" Other causes of measured variation in a trait are characterized as environmental factors, including observational error. In human studies of heritability these are often apportioned into factors from "shared environment" and "non-shared environment" based on whether they tend to result in persons brought up in the same household being more or less similar to persons who were not. Heritability is estimated by comparing individual phenotypic variation among related individuals in a population, by examining the association between ind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
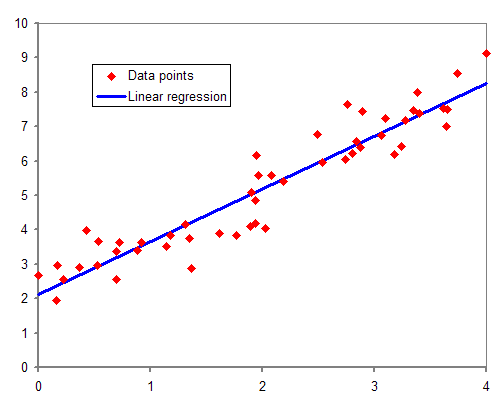
Regression Analysis
In statistical modeling, regression analysis is a set of statistical processes for estimating the relationships between a dependent variable (often called the 'outcome' or 'response' variable, or a 'label' in machine learning parlance) and one or more independent variables (often called 'predictors', 'covariates', 'explanatory variables' or 'features'). The most common form of regression analysis is linear regression, in which one finds the line (or a more complex linear combination) that most closely fits the data according to a specific mathematical criterion. For example, the method of ordinary least squares computes the unique line (or hyperplane) that minimizes the sum of squared differences between the true data and that line (or hyperplane). For specific mathematical reasons (see linear regression), this allows the researcher to estimate the conditional expectation (or population average value) of the dependent variable when the independent variables take on a given ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Marker
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed. A genetic marker may be a short DNA sequence, such as a sequence surrounding a single base-pair change ( single nucleotide polymorphism, SNP), or a long one, like minisatellites. Background For many years, gene mapping was limited to identifying organisms by traditional phenotypes markers. This included genes that encoded easily observable characteristics such as blood types or seed shapes. The insufficient number of these types of characteristics in several organisms limited the mapping efforts that could be done. This prompted the development of gene markers which could identify genetic characteristics that are not readily observable in organisms (such as protein variation). Types Some commonly used types of genetic markers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Linkage
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor Mendel's Law of Independent Assortment. The first experiment to demonstrate li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Null Hypothesis
In scientific research, the null hypothesis (often denoted ''H''0) is the claim that no difference or relationship exists between two sets of data or variables being analyzed. The null hypothesis is that any experimentally observed difference is due to chance alone, and an underlying causative relationship does not exist, hence the term "null". In addition to the null hypothesis, an alternative hypothesis is also developed, which claims that a relationship does exist between two variables. Basic definitions The ''null hypothesis'' and the ''alternative hypothesis'' are types of conjectures used in statistical tests, which are formal methods of reaching conclusions or making decisions on the basis of data. The hypotheses are conjectures about a statistical model of the population, which are based on a sample of the population. The tests are core elements of statistical inference, heavily used in the interpretation of scientific experimental data, to separate scientific claims fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chi-square Statistic
Pearson's chi-squared test (\chi^2) is a statistical test applied to sets of categorical data to evaluate how likely it is that any observed difference between the sets arose by chance. It is the most widely used of many chi-squared tests (e.g., Yates, likelihood ratio, portmanteau test in time series, etc.) – statistical procedures whose results are evaluated by reference to the chi-squared distribution. Its properties were first investigated by Karl Pearson in 1900. In contexts where it is important to improve a distinction between the test statistic and its distribution, names similar to ''Pearson χ-squared'' test or statistic are used. It tests a null hypothesis stating that the frequency distribution of certain events observed in a sample is consistent with a particular theoretical distribution. The events considered must be mutually exclusive and have total probability 1. A common case for this is where the events each cover an outcome of a categorical variable. A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code, or its genotype, and the influence of environmental factors. Both factors may interact, further affecting phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book ''The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as cad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Correlation
In multivariate quantitative genetics, a genetic correlation (denoted r_g or r_a) is the proportion of variance that two traits share due to genetic causes, the correlation between the genetic influences on a trait and the genetic influences on a different trait Plomin et al., p. 123 estimating the degree of pleiotropy or causal overlap. A genetic correlation of 0 implies that the genetic effects on one trait are independent of the other, while a correlation of 1 implies that all of the genetic influences on the two traits are identical. The bivariate genetic correlation can be generalized to inferring genetic latent variable factors across > 2 traits using factor analysis. Genetic correlation models were introduced into behavioral genetics in the 1970s–1980s. Genetic correlations have applications in validation of genome-wide association study (GWAS) results, breeding, prediction of traits, and discovering the etiology of traits & diseases. They can be estimated using indivi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coefficient Of Determination
In statistics, the coefficient of determination, denoted ''R''2 or ''r''2 and pronounced "R squared", is the proportion of the variation in the dependent variable that is predictable from the independent variable(s). It is a statistic used in the context of statistical models whose main purpose is either the prediction of future outcomes or the testing of hypotheses, on the basis of other related information. It provides a measure of how well observed outcomes are replicated by the model, based on the proportion of total variation of outcomes explained by the model. There are several definitions of ''R''2 that are only sometimes equivalent. One class of such cases includes that of simple linear regression where ''r''2 is used instead of ''R''2. When only an intercept is included, then ''r''2 is simply the square of the sample correlation coefficient (i.e., ''r'') between the observed outcomes and the observed predictor values. If additional regressors are included, ''R''2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polygenic
A polygene is a member of a group of non-epistatic genes that interact additively to influence a phenotypic trait, thus contributing to multiple-gene inheritance (polygenic inheritance, multigenic inheritance, quantitative inheritance), a type of non-Mendelian inheritance, as opposed to single-gene inheritance, which is the core notion of Mendelian inheritance. The term "monozygous" is usually used to refer to a hypothetical gene as it is often difficult to distinguish the effect of an individual gene from the effects of other genes and the environment on a particular phenotype. Advances in statistical methodology and high throughput sequencing are, however, allowing researchers to locate candidate genes for the trait. In the case that such a gene is identified, it is referred to as a quantitative trait locus (QTL). These genes are generally pleiotropic as well. The genes that contribute to type 2 diabetes are thought to be mostly polygenes. In July 2016, scientists reported identif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with inc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |