|
Lead Finder
Lead Finder software is a computational chemistry application for modeling protein-ligand interactions. The software can be used in molecular docking studies, for the quantitative evaluation of ligand binding and biological activity. For individual, non-commercial and academic users the software is free. About Lead Finder software is an integrated solution for simulating structure and binding affinity of protein-ligand complexes. The software combines automatic processing of protein structures, extra precision protein-ligand docking and calculation of free energy of ligand binding. Original docking algorithm provides a fast rate of calculations, which can be easily adjusted from more rapid (for virtual screening applications) to slightly more slow and robust, while unique scoring function implemented in Lead Finder provides unsurpassed accuracy of calculations. Lead Finder is intended to meet the requirements of computational and medicinal chemists involved in drug discovery, ph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Computational Chemistry
Computational chemistry is a branch of chemistry that uses computer simulation to assist in solving chemical problems. It uses methods of theoretical chemistry, incorporated into computer programs, to calculate the structures and properties of molecules, groups of molecules, and solids. It is essential because, apart from relatively recent results concerning the hydrogen molecular ion ( dihydrogen cation, see references therein for more details), the quantum many-body problem cannot be solved analytically, much less in closed form. While computational results normally complement the information obtained by chemical experiments, it can in some cases predict hitherto unobserved chemical phenomena. It is widely used in the design of new drugs and materials. Examples of such properties are structure (i.e., the expected positions of the constituent atoms), absolute and relative (interaction) energies, electronic charge density distributions, dipoles and higher multipole moments ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Molecular Docking
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions. The associations between biologically relevant molecules such as proteins, peptides, nucleic acids, carbohydrates, and lipids play a central role in signal transduction. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced. Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target bindi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Biological Activity
In pharmacology, biological activity or pharmacological activity describes the beneficial or adverse effects of a drug on living matter. When a drug is a complex chemical mixture, this activity is exerted by the substance's active ingredient or pharmacophore but can be modified by the other constituents. Among the various properties of chemical compounds, pharmacological/biological activity plays a crucial role since it suggests uses of the compounds in the medical applications. However, chemical compounds may show some adverse and toxic effects which may prevent their use in medical practice. Activity is generally dosage-dependent. Further, it is common to have effects ranging from beneficial to adverse for one substance when going from low to high doses. Activity depends critically on fulfillment of the ADME criteria. To be an effective drug, a compound not only must be active against a target, but also possess the appropriate ADME (Absorption, Distribution, Metabolism, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Binding Affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of lig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
 |
Virtual Screening
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor or enzyme. Virtual screening has been defined as "automatically evaluating very large libraries of compounds" using computer programs. As this definition suggests, VS has largely been a numbers game focusing on how the enormous chemical space of over 1060 conceivable compounds can be filtered to a manageable number that can be synthesized, purchased, and tested. Although searching the entire chemical universe may be a theoretically interesting problem, more practical VS scenarios focus on designing and optimizing targeted combinatorial libraries and enriching libraries of available compounds from in-house compound repositories or vendor offerings. As the accuracy of the method has increased, virtual screening has become an integral part of the drug disc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Drug Discovery
In the fields of medicine, biotechnology and pharmacology, drug discovery is the process by which new candidate medications are discovered. Historically, drugs were discovered by identifying the active ingredient from traditional remedies or by serendipitous discovery, as with penicillin. More recently, chemical libraries of synthetic small molecules, natural products or extracts were screened in intact cells or whole organisms to identify substances that had a desirable therapeutic effect in a process known as classical pharmacology. After sequencing of the human genome allowed rapid cloning and synthesis of large quantities of purified proteins, it has become common practice to use high throughput screening of large compounds libraries against isolated biological targets which are hypothesized to be disease-modifying in a process known as reverse pharmacology. Hits from these screens are then tested in cells and then in animals for efficacy. Modern drug discovery i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
ADME-Tox
ADME is an abbreviation in pharmacokinetics and pharmacology for "absorption (pharmacokinetics), absorption, distribution (pharmacology), distribution, metabolism, and excretion", and describes the Drug Disposition, disposition of a pharmaceutical Chemical compound, compound within an organism. The four criteria all influence the drug metabolism, drug levels and kinetics of drug exposure to the tissues and hence influence the performance and pharmacological activity of the compound as a drug. Sometimes, liberation (pharmacology), liberation and/or toxicity are also considered, yielding LADME, ADMET, or LADMET. Components Absorption/administration For a compound to reach a tissue, it usually must be taken into the bloodstream – often via mucus, mucous surfaces like the digestive tract (intestinal absorption) – before being taken up by the target cells. Factors such as poor compound solubility, gastric emptying time, intestinal transit time, chemical instability in the st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
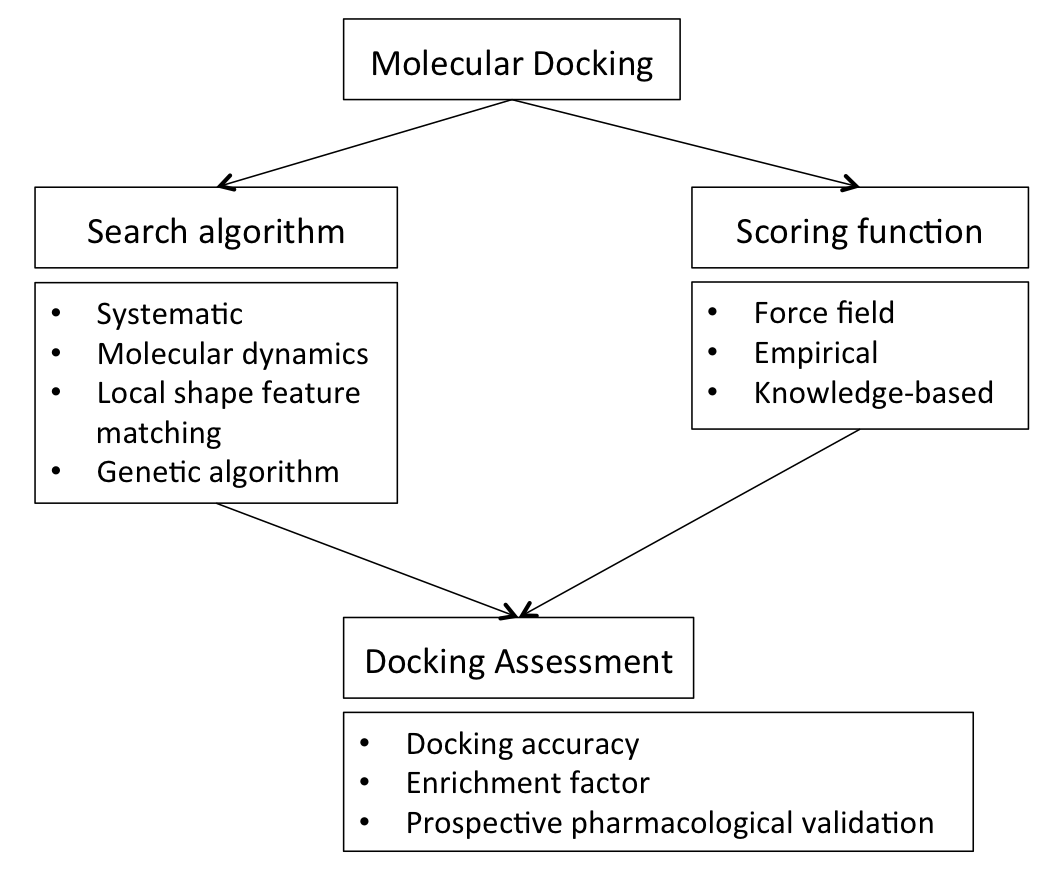 |
Ligand Docking
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions. The associations between biologically relevant molecules such as proteins, peptides, nucleic acids, carbohydrates, and lipids play a central role in signal transduction. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced. Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target bind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Global Minimum
In mathematical analysis, the maxima and minima (the respective plurals of maximum and minimum) of a function, known collectively as extrema (the plural of extremum), are the largest and smallest value of the function, either within a given range (the ''local'' or ''relative'' extrema), or on the entire domain (the ''global'' or ''absolute'' extrema). Pierre de Fermat was one of the first mathematicians to propose a general technique, adequality, for finding the maxima and minima of functions. As defined in set theory, the maximum and minimum of a set are the greatest and least elements in the set, respectively. Unbounded infinite sets, such as the set of real numbers, have no minimum or maximum. Definition A real-valued function ''f'' defined on a domain ''X'' has a global (or absolute) maximum point at ''x''∗, if for all ''x'' in ''X''. Similarly, the function has a global (or absolute) minimum point at ''x''∗, if for all ''x'' in ''X''. The value of the function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Genetic Algorithm
In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems by relying on biologically inspired operators such as mutation, crossover and selection. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, etc. Methodology Optimization problems In a genetic algorithm, a population of candidate solutions (called individuals, creatures, organisms, or phenotypes) to an optimization problem is evolved toward better solutions. Each candidate solution has a set of properties (its chromosomes or genotype) which can be mutated and altered; traditionally, solutions are represented in binary as strings of 0s and 1s, but other encodings are also po ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Scoring Functions For Docking
In the fields of computational chemistry and molecular modelling, scoring functions are mathematical functions used to approximately predict the binding affinity between two molecules after they have been docked. Most commonly one of the molecules is a small organic compound such as a drug and the second is the drug's biological target such as a protein receptor. Scoring functions have also been developed to predict the strength of intermolecular interactions between two proteins or between protein and DNA. Utility Scoring functions are widely used in drug discovery and other molecular modelling applications. These include: * Virtual screening of small molecule databases of candidate ligands to identify novel small molecules that bind to a protein target of interest and therefore are useful starting points for drug discovery * De novo design (design "from scratch") of novel small molecules that bind to a protein target * Lead optimization of screening hits to optimize ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |