|
Kozak Consensus Sequence
The Kozak consensus sequence (Kozak consensus or Kozak sequence) is a nucleic acid motif that functions as the protein translation initiation site in most eukaryotic mRNA transcripts. Regarded as the optimum sequence for initiating translation in eukaryotes, the sequence is an integral aspect of protein regulation and overall cellular health as well as having implications in human disease. It ensures that a protein is correctly translated from the genetic message, mediating ribosome assembly and translation initiation. A wrong start site can result in non-functional proteins. As it has become more studied, expansions of the nucleotide sequence, bases of importance, and notable exceptions have arisen. The sequence was named after the scientist who discovered it, Marilyn Kozak. Kozak discovered the sequence through a detailed analysis of DNA genomic sequences. The Kozak sequence is not to be confused with the ribosomal binding site (RBS), that being either the 5′ cap of a me ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Sequence
A nucleic acid sequence is a succession of Nucleobase, bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. By convention, sequences are usually presented from the Directionality (molecular biology), 5' end to the 3' end. For DNA, the Sense (molecular biology), sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the Biomolecular structure#Primary structure, primary structure. The sequence has capacity to represent information. Biological deoxyribonucleic acid represents the information which directs the functions of an organism. Nucleic acids also have a Nucleic acid secondary structure, secondary structure and Nucleic acid tertiary structure, tertiary structure. Primary structur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
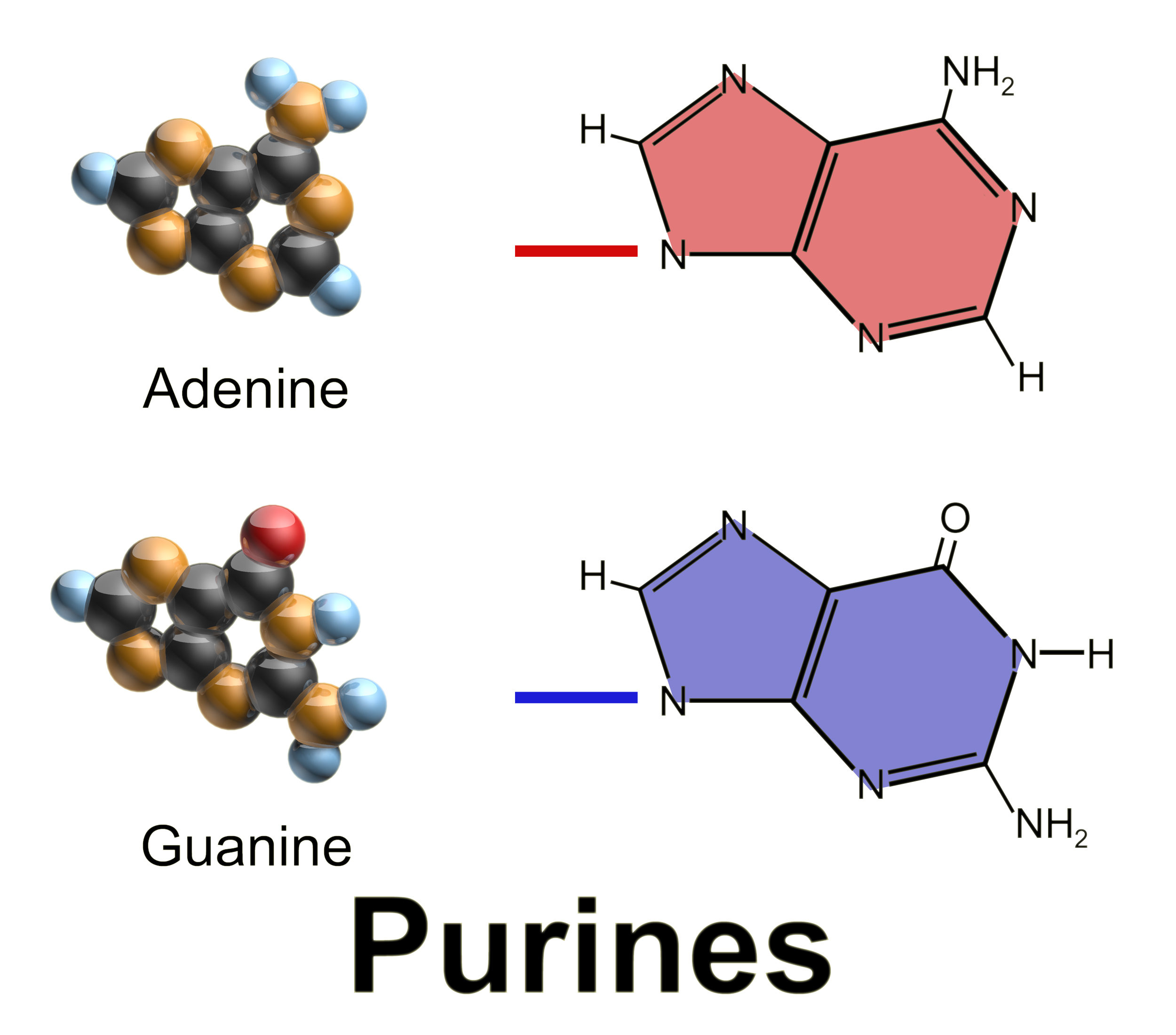
Purine
Purine is a heterocyclic aromatic organic compound that consists of two rings ( pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature. Dietary sources Purines are found in high concentration in meat and meat products, especially internal organs such as liver and kidney. In general, plant-based diets are low in purines. High-purine plants and algae include some legumes (lentils and black eye peas) and spirulina. Examples of high-purine sources include: sweetbreads, anchovies, sardines, liver, beef kidneys, brains, meat extracts (e.g., Oxo, Bovril), herring, mackerel, scallops, game meats, yeast (beer, yeast extract, nutritional yeast) and gravy. A moderate amount of purine is also contained in red meat, beef, pork, poultry, fish and seafood, asparagus, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DDX3X
ATP-dependent RNA helicase DDX3X is an enzyme that in humans is encoded by the ''DDX3X'' gene. Function DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which interacts specifically with hepatitis C virus core protein resulting a change in intracellular location. This gene has a homolog located in the nonrecombining region of the Y chromosome. The protein sequence is 91% identical between this gene and the Y-linked homolog. Sub-cellular trafficking DDX3X performs its functions in the cell nucleus and cytoplasm, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DHX29
DExH-box helicase 29 (DHX29) is a 155 kDa protein that in humans is encoded by the DHX29 gene. Function This gene encodes a member of the DEAH (Asp-Glu-Ala-His) subfamily of proteins, part of the DEAD (Asp-Glu-Ala-Asp) box family of RNA helicases. The encoded protein functions in translation initiation, and is specifically required for ribosomal scanning across stable mRNA secondary structures during initiation codon selection. This protein may also play a role in sensing virally derived cytosolic nucleic acids. Knockdown of this gene results in reduced protein translation and impaired proliferation of cancer cells. Interactions DHX29 has been shown to interact with the eukaryotic small ribosomal subunit (40S) The eukaryotic small ribosomal subunit (40S) is the smaller subunit of the eukaryotic 80S ribosomes, with the other major component being the large ribosomal subunit (60S). The "40S" and "60S" names originate from the convention that ribosomal pa ... and eI ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Initiation
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Eukar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
7-Methylguanosine
7-Methylguanosine (m7G) is a modified purine nucleoside. It is a methylated version of guanosine and when found in human urine, it may be a biomarker of some types of cancer. In the RNAs, 7-methylguanosine have been used to study and examine the reaction evolving methylguanosine. It also plays a role in mRNA as a blocking group at its 5´-end. See also * METTL1 *23S rRNA (guanine2445-N2)-methyltransferase *16S rRNA (guanine527-N7)-methyltransferase 16S rRNA (guanine527-N7)-methyltransferase (, ''ribosomal RNA small subunit methyltransferase G'', ''16S rRNA methyltransferase RsmG'', ''GidB'', ''rsmG (gene)'') is an enzyme with systematic name ''S-adenosyl-L-methionine:16S rRNA (guanine527-N7)- ... References External links Metabocard for 7-Methylguanosine (HMDB01107) Human Metabolome Database, University of Alberta Nucleosides Purines Hydroxymethyl compounds Biomarkers {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PABPC1
Polyadenylate-binding protein 1 is a protein that in humans is encoded by the ''PABPC1'' gene. The protein PABP1 binds mRNA and facilitates a variety of functions such as transport into and out of the nucleus, degradation, translation, and stability. There are two separate PABP1 proteins, one which is located in the nucleus (PABPN1) and the other which is found in the cytoplasm (PABPC1). The location of PABP1 affects the role of that protein and its function with RNA. Function The poly(A)-binding protein (PAB or PABP), which is found complexed to the 3' poly(A) tail of eukaryotic mRNA, is required for poly(A) lengthening and the termination of translation. In humans, the PABPs comprise a small nuclear isoform and a conserved gene family of other poly(A)-binding proteins. upplied by OMIMref name="entrez"> PABPC1 is usually diffused within the cytoplasm and concentrated at sites of high mRNA concentration such as stress granules, processing bodies, and locations of high translat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Initiation Factor 3
Eukaryotic initiation factor 3 (eIF3) is a multiprotein complex that functions during the initiation phase of eukaryotic translation. It is essential for most forms of cap-dependent and cap-independent translation initiation. In humans, eIF3 consists of 13 nonidentical subunits (eIF3a-m) with a combined molecular weight of ~800 kDa, making it the largest translation initiation factor. The eIF3 complex is broadly conserved across eukaryotes, but the conservation of individual subunits varies across organisms. For instance, while most mammalian eIF3 complexes are composed of 13 subunits, budding yeast's eIF3 has only six subunits (eIF3a, b, c, g, i, j). Function eIF3 stimulates nearly all steps of translation initiation. eIF3 also appears to participate in other phases of translation, such as recycling, where it promotes the splitting of post-termination ribosomes. In specialized cases of reinitiation following uORFs, eIF3 may remain bound to the ribosome through elongation a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EIF5
Eukaryotic translation initiation factor 5 is a protein that in humans is encoded by the ''EIF5'' gene. EIF5 is a GTPase-activating protein GTPase-activating proteins or GTPase-accelerating proteins (GAPs) are a family of regulatory proteins whose members can bind to activated G proteins and stimulate their GTPase activity, with the result of terminating the signaling event. GAPs are a .... References External links Cap-dependent translation initiationfrom Nature Reviews Microbiology. A good image and overview of the function of initiation factors PDBe-KBprovides an overview of all the structure information available in the PDB for Human Eukaryotic translation initiation factor 5 (EIF5) Further reading * * * * * * * * * * * * * * * * * {{gene-14-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EIF2
Eukaryotic Initiation Factor 2 (eIF2) is an eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha (also called subunit 1, EIF2S1), a beta (subunit 2, EIF2S2), and a gamma (subunit 3, EIF2S3) subunit. Once the initiation phase has completed, eIF2 is released from the ribosome bound to GDP as an inactive binary complex. To participate in another round of translation initiation, this GDP must be exchanged for GTP. Function eIF2 is an essential factor for protein synthesis that forms a ternary complex (TC) with GTP and the initiator Met-tRNAiMet. After its formation, the TC binds the 40S ribosomal subunit to form the 43S preinitiation complex (43S PIC). 43S PIC assembly is believed to be stimulated by the initiation factors eIF1, eIF1A, and the eIF3 complex according to ''in vitro'' experiments. The 43S P ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA, transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain vi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |