|
Jun Dimerization Protein
Jun dimerization protein 2 (JUNDM2) is a protein that in humans is encoded by the ''JDP2'' gene. The Jun dimerization protein is a member of the AP-1 family of transcription factors. JDP 2 was found by a Sos-recruitment system, to dimerize with c-Jun to repress AP-1-mediated activation. It was later identified by the yeast-two hybrid system to bind to activating transcription factor 2 (ATF2) to repress ATF-mediated transcriptional activation. JDP2 regulates 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE)- and cAMP-responsive element (CRE)-dependent transcription. The JDP2 gene is located on human chromosome 14q24.3 (46.4 kb, 75,427,715 bp to 75,474,111 bp) and mouse chromosome 12 (39 kb, 85,599,105 bp to 85,639,878 bp), which is located at about 250 kbp in the Fos-JDP2-BATF locus. Alternative splicing of JDP2 generates at least two isoforms. The protein JDP2 has 163 amino acids, belongs to the family of basic leucine zipper (bZIP), and shows high homology ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid resid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
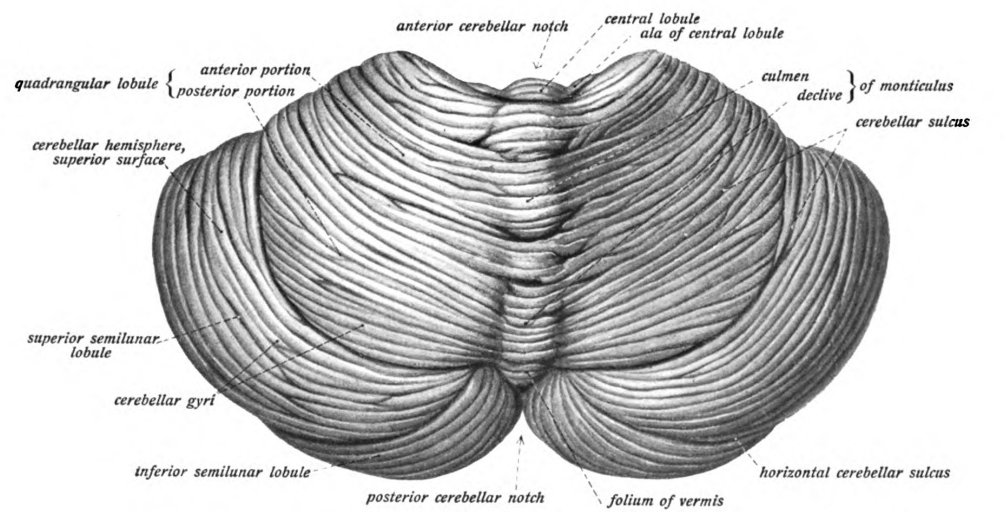
Cerebellum
The cerebellum (Latin for "little brain") is a major feature of the hindbrain of all vertebrates. Although usually smaller than the cerebrum, in some animals such as the mormyrid fishes it may be as large as or even larger. In humans, the cerebellum plays an important role in motor control. It may also be involved in some cognitive functions such as attention and language as well as emotional control such as regulating fear and pleasure responses, but its movement-related functions are the most solidly established. The human cerebellum does not initiate movement, but contributes to coordination, precision, and accurate timing: it receives input from sensory systems of the spinal cord and from other parts of the brain, and integrates these inputs to fine-tune motor activity. Cerebellar damage produces disorders in fine movement, equilibrium, posture, and motor learning in humans. Anatomically, the human cerebellum has the appearance of a separate structure attached to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HDAC2
Histone deacetylase 2 (HDAC2) is an enzyme that in humans is encoded by the ''HDAC2'' gene. It belongs to the histone deacetylase class of enzymes responsible for the removal of acetyl groups from lysine residues at the N-terminal region of the core histones (H2A, H2B, H3, and H4). As such, it plays an important role in gene expression by facilitating the formation of transcription repressor complexes and for this reason is often considered an important target for cancer therapy. Though the functional role of the class to which HDAC2 belongs has been carefully studied, the mechanism by which HDAC2 interacts with histone deacetylases of other classes has yet to be elucidated. HDAC2 is broadly regulated by protein kinase 2 (CK2) and protein phosphatase 1 (PP1), but biochemical analysis suggests its regulation is more complex (evinced by the coexistence of HDAC1 and HDAC2 in three distinct protein complexes). Essentially, the mechanism by which HDAC2 is regulated is still unclear by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HDAC1
Histone deacetylase 1 (HDAC1) is an enzyme that in humans is encoded by the ''HDAC1'' gene. Function Histone acetylation and deacetylation, catalyzed by multisubunit complexes, play a key role in the regulation of eukaryotic gene expression. The protein encoded by this gene belongs to the histone deacetylase/acuc/apha family and is a component of the histone deacetylase complex. It also interacts with retinoblastoma tumor-suppressor protein and this complex is a key element in the control of cell proliferation and differentiation. Together with metastasis-associated protein-2 MTA2, it deacetylates p53 and modulates its effect on cell growth and apoptosis. Model organisms Model organisms have been used in the study of HDAC1 function. A conditional knockout mouse line, called ''Hdac1tm1a(EUCOMM)Wtsi'' was generated as part of the International Knockout Mouse Consortium program — a high-throughput mutagenesis project to generate and distribute animal models of disease to in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IRF2BP1
IRF may refer to: General *Impulse response function *Information Retrieval Facility * Initial Reaction Force or Internal Response Force * Immediate Response Force *Institute of Space Physics (Sweden), (Institutet för rymdfysik) *Interferon Regulatory Factor (e.g. IRF6) *International Rectifier, New York Stock Exchange symbol IRF Foundations/organizations *International Ranger Federation *International Rabbinic Fellowship * International Rogaining Federation *Islamic Research Foundation *International Rafting Federation *International Ringette Federation The International Ringette Federation (IRF) is a non-profit amateur sports organization and the highest governing body for the sport of ringette. Today the member countries of the IRF Board includes four member nations: Canada, Finland, Sweden, a ... Computing * Intelligent Resilient Framework, Virtual Switch Chassis Aggregation {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain lysyl ((CH2)4NH2), classifying it as a basic, charged (at physiological pH), aliphatic amino acid. It is encoded by the codons AAA and AAG. Like almost all other amino acids, the α-carbon is chiral and lysine may refer to either enantiomer or a racemic mixture of both. For the purpose of this article, lysine will refer to the biologically active enantiomer L-lysine, where the α-carbon is in the ''S'' configuration. The human body cannot synthesize lysine. It is essential in humans and must therefore be obtained from the diet. In organisms that synthesise lysine, two main biosynthetic pathways exist, the diaminopimelate and α-aminoadipate pathways, which employ distinct ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SUMO Protein
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes written sumoylation). SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. SUMO proteins are similar to ubiquitin and are considered members of the ubiquitin-like protein family. SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO is not used to tag proteins for degradation. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P38 Mitogen-activated Protein Kinases
p38 mitogen-activated protein kinases are a class of mitogen-activated protein kinases (MAPKs) that are responsive to stress stimuli, such as cytokines, ultraviolet irradiation, heat shock, and osmotic shock, and are involved in cell differentiation, apoptosis and autophagy. Persistent activation of the p38 MAPK pathway in muscle satellite cells (muscle stem cells) due to ageing, impairs muscle regeneration. p38 MAP Kinase (MAPK), also called RK or CSBP (Cytokinin Specific Binding Protein), is the mammalian orthologue of the yeast Hog1p MAP kinase, which participates in a signaling cascade controlling cellular responses to cytokines and stress. Four p38 MAP kinases, p38-α ( MAPK14), -β ( MAPK11), -γ (MAPK12 / ERK6), and -δ (MAPK13 / SAPK4), have been identified. Similar to the SAPK/JNK pathway, p38 MAP kinase is activated by a variety of cellular stresses including osmotic shock, inflammatory cytokines, lipopolysaccharides (LPS), ultraviolet light, and growth facto ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
JNK1
Mitogen-activated protein kinase 8 (also known as JNK1) is a ubiquitous enzyme that in humans is encoded by the ''MAPK8'' gene. Function The protein encoded by this gene is a member of the MAP kinase and JNK family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. This kinase is activated by various cell stimuli, and targets specific transcription factors, and thus mediates immediate-early gene expression in response to cell stimuli. The activation of this kinase by tumor-necrosis factor alpha (TNF-alpha) is found to be required for TNF-alpha-induced apoptosis. This kinase is also involved in UV radiation-induced apoptosis, which is thought to be related to the cytochrome c-mediated cell death pathway. Studies of the mouse counterpart of this gene suggested that this kinase play a key role in T cell proliferation, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAPK8
Mitogen-activated protein kinase 8 (also known as JNK1) is a ubiquitous enzyme that in humans is encoded by the ''MAPK8'' gene. Function The protein encoded by this gene is a member of the Mitogen-activated protein kinase, MAP kinase and c-Jun N-terminal kinases, JNK family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as Cell proliferation, proliferation, Cellular differentiation, differentiation, transcription regulation and development. This kinase is activated by various cell stimuli, and targets specific transcription factors, and thus mediates immediate-early gene expression in response to cell stimuli. The activation of this kinase by Tumor necrosis factor-alpha, tumor-necrosis factor alpha (TNF-alpha) is found to be required for TNF-alpha-induced apoptosis. This kinase is also involved in UV radiation-induced apoptosis, which is thought to be related to the cytochrome c-mediated ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), a carboxyl group (which is in the deprotonated −COO− form under biological conditions), and a side chain containing a hydroxyl group, making it a polar, uncharged amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Threonine is synthesized from aspartate in bacteria such as ''E. coli''. It is encoded by all the codons starting AC (ACU, ACC, ACA, and ACG). Threonine sidechains are often hydrogen bonded; the most common small motifs formed are based on interactions with serine: ST turns, ST motifs (often at the beginning of alpha helices) and ST staples (usually at the middle of alpha helices). Modifications The threonine residue is susceptible to numerous posttranslational modifications. The hydroxyl side-chain can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |