|
Intramembrane Protease
Intramembrane proteases (IMPs), also known as intramembrane-cleaving proteases (I-CLiPs), are enzymes that have the property of cleaving transmembrane domains of integral membrane proteins. All known intramembrane proteases are themselves integral membrane proteins with multiple transmembrane domains, and they have their active sites buried within the lipid bilayer of cellular membranes. Intramembrane proteases are responsible for proteolytic cleavage in the cell signaling process known as regulated intramembrane proteolysis (RIP). Intramembrane proteases are not evolutionarily related to classical soluble proteases, having evolved their catalytic sites by convergent evolution. Although only recently discovered, intramembrane proteases are of significant research interest because of their major biological functions and their relevance to human disease. Classification There are four groups of intramembrane proteases, distinguished by their catalytic mechanism: *Metalloproteases: ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Rhomboid Protease
The rhomboid proteases are a family of enzymes that exist in almost all species. They are proteases: they cut the polypeptide chain of other proteins. This proteolytic cleavage is irreversible in cells, and an important type of cellular regulation. Although proteases are one of the earliest and best studied class of enzyme, rhomboids belong to a much more recently discovered type: the intramembrane proteases. What is unique about intramembrane proteases is that their active sites are buried in the lipid bilayer of cell membranes, and they cleave other transmembrane proteins within their transmembrane domains. About 30% of all proteins have transmembrane domains, and their regulated processing often has major biological consequences. Accordingly, rhomboids regulate many important cellular processes, and may be involved in a wide range of human diseases. Intramembrane proteases Rhomboids are intramembrane serine proteases. The other types of intramembrane protease are aspartyl- and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Scissile Bond
In molecular biology, a scissile bond is a covalent chemical bond that can be broken by an enzyme. Examples would be the cleaved bond in the self-cleaving hammerhead ribozyme or the peptide bond In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 ( nitrogen number two) of another, along a peptide or protein c ... of a substrate cleaved by a peptidase. References {{Reflist Enzymes Molecular biology Chemical bonding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Cryo-electron Microscopy
Cryogenic electron microscopy (cryo-EM) is a cryomicroscopy technique applied on samples cooled to cryogenic temperatures. For biological specimens, the structure is preserved by embedding in an environment of vitreous ice. An aqueous sample solution is applied to a grid-mesh and plunge-frozen in liquid ethane or a mixture of liquid ethane and propane. While development of the technique began in the 1970s, recent advances in detector technology and software algorithms have allowed for the determination of biomolecular structures at near-atomic resolution. This has attracted wide attention to the approach as an alternative to X-ray crystallography or NMR spectroscopy for macromolecular structure determination without the need for crystallization. In 2017, the Nobel Prize in Chemistry was awarded to Jacques Dubochet, Joachim Frank, and Richard Henderson "for developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution." ''Nature ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
 |
X-ray Crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information. Since many materials can form crystals—such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules—X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Gamma-secretase
Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44. Subunits and assembly The gamma secretase complex consists of four individual proteins: PSEN1 (presenilin-1), nicastrin, APH-1 (anterior pharynx-defective 1), and PEN-2 (presenilin enhancer 2). Rece ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
 |
Hydrophobic
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exception ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
Aqueous
An aqueous solution is a solution in which the solvent is water. It is mostly shown in chemical equations by appending (aq) to the relevant chemical formula. For example, a solution of table salt, or sodium chloride (NaCl), in water would be represented as . The word ''aqueous'' (which comes from ''aqua'') means pertaining to, related to, similar to, or dissolved in, water. As water is an excellent solvent and is also naturally abundant, it is a ubiquitous solvent in chemistry. Since water is frequently used as the solvent in experiments, the word solution refers to an aqueous solution, unless the solvent is specified. A ''non-aqueous solution'' is a solution in which the solvent is a liquid, but is not water. (See also Solvent and Inorganic nonaqueous solvent.) Characteristics Substances that are '' hydrophobic'' ('water-fearing') do not dissolve well in water, whereas those that are '' hydrophilic'' ('water-friendly') do. An example of a hydrophilic substance is sod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
|
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate ( binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
|
.png) |
Transmembrane Helices
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
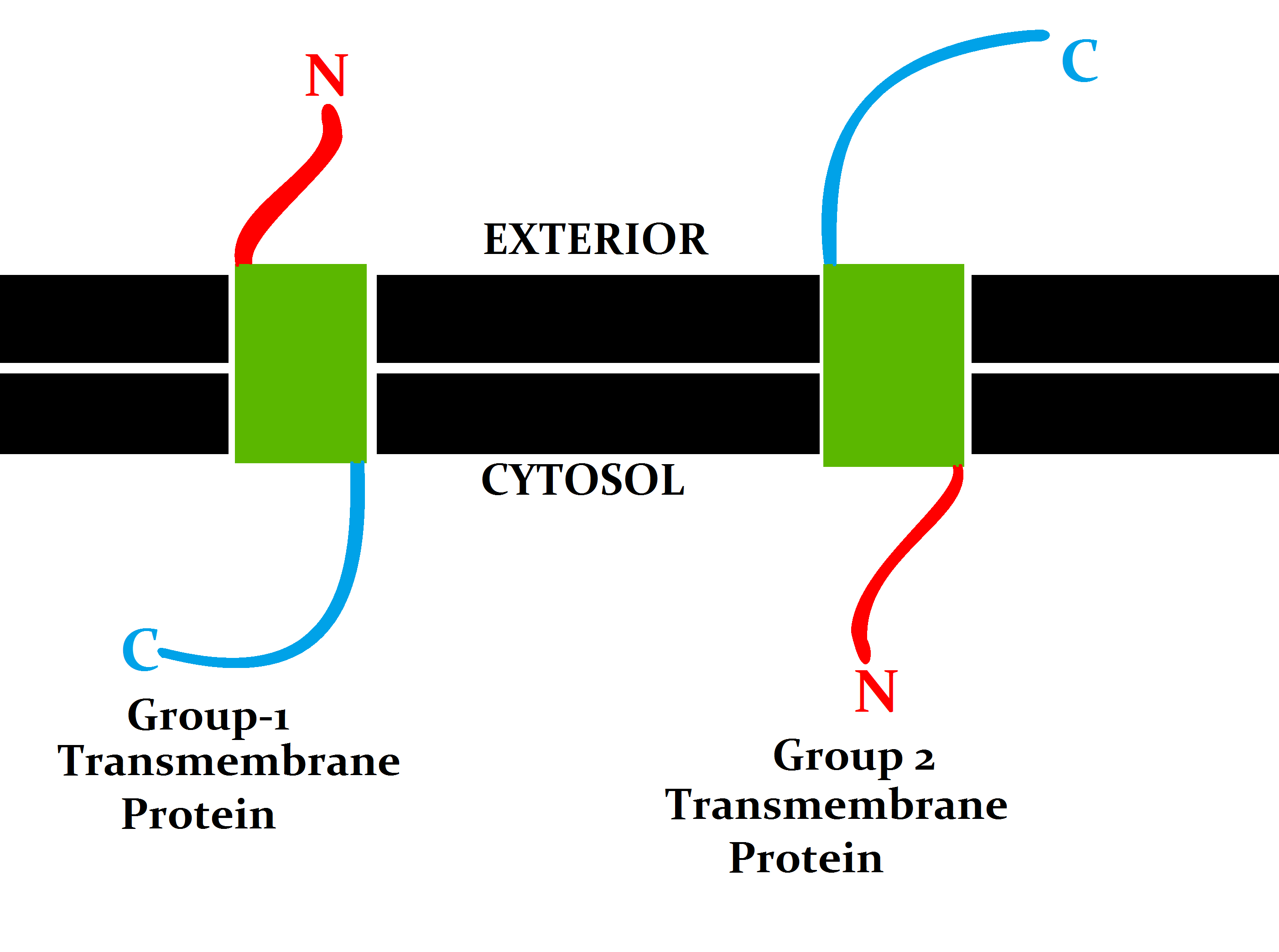 |
Transmembrane Protein
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them ( beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |