|
Integrative Bioinformatics
Integrative bioinformatics is a discipline of bioinformatics that focuses on problems of data integration for the life sciences. With the rise of high-throughput (HTP) technologies in the life sciences, particularly in molecular biology, the amount of collected data has grown in an exponential fashion. Furthermore, the data are scattered over a plethora of both public and private repositories, and are stored using a large number of different formats. This situation makes searching these data and performing the analysis necessary for the extraction of new knowledge from the complete set of available data very difficult. Integrative bioinformatics attempts to tackle this problem by providing unified access to life science data. Approaches Semantic web approaches In the Semantic Web approach, data from multiple websites or databases is searched via metadata. Metadata is machine-readable code, which defines the contents of the page for the program so that the comparisons between ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms ( SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable propertie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PubMed
PubMed is a free search engine accessing primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine (NLM) at the National Institutes of Health maintain the database as part of the Entrez system of information retrieval. From 1971 to 1997, online access to the MEDLINE database had been primarily through institutional facilities, such as university libraries. PubMed, first released in January 1996, ushered in the era of private, free, home- and office-based MEDLINE searching. The PubMed system was offered free to the public starting in June 1997. Content In addition to MEDLINE, PubMed provides access to: * older references from the print version of '' Index Medicus'', back to 1951 and earlier * references to some journals before they were indexed in Index Medicus and MEDLINE, for instance ''Science'', '' BMJ'', and '' Annals of Surgery'' * very recent entries to records for an article ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Data Visualization
Biology data visualization is a branch of bioinformatics concerned with the application of computer graphics, scientific visualization, and information visualization to different areas of the life sciences. This includes visualization of sequences, genomes, alignments, phylogenies, macromolecular structures, systems biology, microscopy, and magnetic resonance imaging data. Software tools used for visualizing biological data range from simple, standalone programs to complex, integrated systems. State-of-the-art and perspectives Today we are experiencing a rapid growth in volume and diversity of biological data, presenting an increasing challenge for biologists. A key step in understanding and learning from these data is visualization. Thus, there has been a corresponding increase in the number and diversity of systems for visualizing biological data. An emerging trend is the blurring of boundaries between the visualization of 3D structures at atomic resolution, visualizatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Database
Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genomics, proteomics, metabolomics, microarray gene expression, and phylogenetics. Information contained in biological databases includes gene function, structure, localization (both cellular and chromosomal), clinical effects of mutations as well as similarities of biological sequences and structures. Biological databases can be classified by the kind of data they collect (see below). Broadly, there are molecular databases (for sequences, molecules, etc.), functional databases (for physiology, enzyme activities, phenotypes, ecology etc), taxonomic databases (for species and other taxonomic ranks), images and other media, or specimens (for museum collections etc.) Databases are important tools in assisting scientists to analyze and explain a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteomics Standards Initiative
The Proteomics Standards Initiative (PSI) is a working group of the Human Proteome Organization. It aims to define data standards for proteomics to facilitate data comparison, exchange and verification. The Proteomics Standards Initiative focuses on the following subjects: minimum information about a proteomics experiment defines the metadata that should be provided along with a proteomics experiment. a data markup language for encoding the data, and metadata ontologies for consistent annotation and representation. Minimum information about a proteomics experiment Minimum information about a proteomics experiment (MIAPE) is a minimum information standard, created by the Proteomics Standards Initiative of the Human Proteome Organization, for reporting proteomics experiments. You can't just introduce the results of an analysis, it is intended to specify all the information necessary to interpret the experiment results unambiguously and to potentially reproduce the experiment. While ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Omics Topics In Biology
Inspired by the terms genome and genomics, other words to describe complete biological datasets, mostly sets of biomolecules originating from one organism, have been coined with the suffix ''-ome'' and ''- omics''. Some of these terms are related to each other in a hierarchical fashion. For example, the genome contains the ORFeome, which gives rise to the transcriptome, which is translated to the proteome. Other terms are overlapping and refer to the structure and/or function of a subset of proteins (e.g. glycome, kinome). An omicist is a scientist who studies omeomics, cataloging all the “omics” subfields. Omics.org is a Wiki that collects and alphabetically lists all the known "omes" and "omics." List of topics Hierarchy of topics For the sake of clarity, some topics are listed more than once. * Bibliome * Connectome * Cytome *Editome *Embryome * Epigenome ** Methylome *Exposome **Envirome *** Toxome ** Foodome **Microbiome ** Sociome *Genome ** Variome ** Exome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
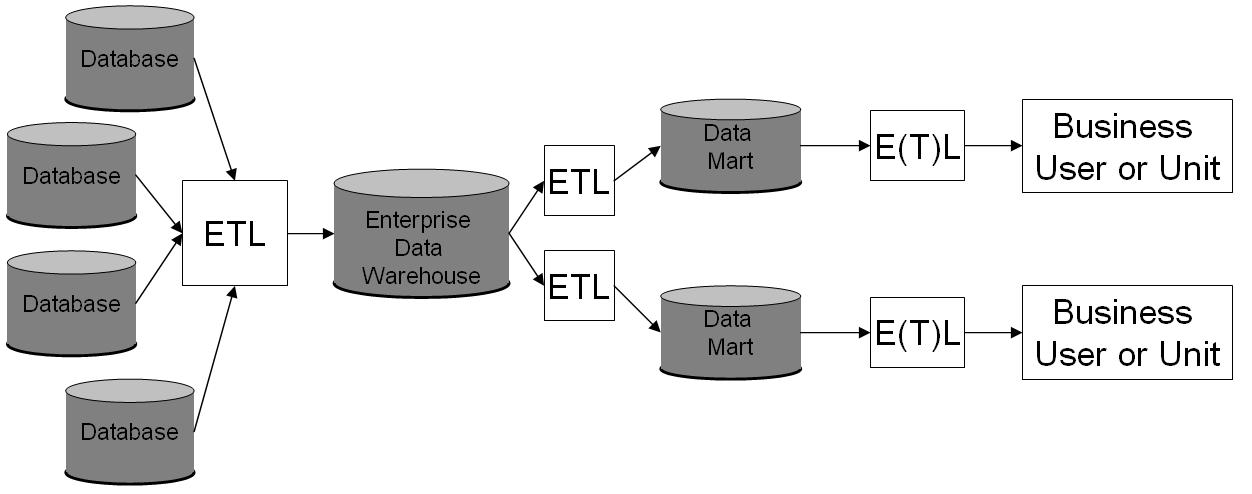
Data Warehousing
In computing, a data warehouse (DW or DWH), also known as an enterprise data warehouse (EDW), is a system used for reporting and data analysis and is considered a core component of business intelligence. DWs are central repositories of integrated data from one or more disparate sources. They store current and historical data in one single place that are used for creating analytical reports for workers throughout the enterprise. The data stored in the warehouse is uploaded from the operational systems (such as marketing or sales). The data may pass through an operational data store and may require data cleansing for additional operations to ensure data quality before it is used in the DW for reporting. Extract, transform, load (ETL) and extract, load, transform (ELT) are the two main approaches used to build a data warehouse system. ETL-based data warehousing The typical extract, transform, load (ETL)-based data warehouse uses staging, data integration, and access layers t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Natural Language Processing
Natural language processing (NLP) is an interdisciplinary subfield of linguistics, computer science, and artificial intelligence concerned with the interactions between computers and human language, in particular how to program computers to process and analyze large amounts of natural language data. The goal is a computer capable of "understanding" the contents of documents, including the contextual nuances of the language within them. The technology can then accurately extract information and insights contained in the documents as well as categorize and organize the documents themselves. Challenges in natural language processing frequently involve speech recognition, natural-language understanding, and natural-language generation. History Natural language processing has its roots in the 1950s. Already in 1950, Alan Turing published an article titled " Computing Machinery and Intelligence" which proposed what is now called the Turing test as a criterion of intelligence, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TrEMBL
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ensembl
Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other vertebrates and model organisms. Ensembl is one of several well known genome browsers for the retrieval of genomic information. Similar databases and browsers are found at NCBI and the University of California, Santa Cruz (UCSC). History The human genome consists of three billion base pairs, which code for approximately 20,000–25,000 genes. However the genome alone is of little use, unless the locations and relationships of individual genes can be identified. One option is manual annotation, whereby a team of scientists tries to locate genes using experimental data from scientific journals and public databases. However this is a slow, painstaking task. The alternative, known as automated annotation, is to use the power of comput ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SwissProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |