|
Influenza Non-structural Protein
Influenza non-structural protein (NS1) is a homodimeric RNA-binding protein found in influenza virus that is required for viral replication. NS1 binds polyA tails of mRNA keeping them in the nucleus. NS1 inhibits pre-mRNA splicing by tightly binding to a specific stem-bulge of U6 snRNA U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a .... References {{DEFAULTSORT:Influenza Non-Structural Protein Protein families ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Influenza Virus
''Orthomyxoviridae'' (from Greek ὀρθός, ''orthós'' 'straight' + μύξα, ''mýxa'' 'mucus') is a family of negative-sense RNA viruses. It includes seven genera: ''Alphainfluenzavirus'', ''Betainfluenzavirus'', '' Gammainfluenzavirus'', '' Deltainfluenzavirus'', ''Isavirus'', ''Thogotovirus'', and ''Quaranjavirus''. The first four genera contain viruses that cause influenza in birds (see also avian influenza) and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates (such as ticks and mosquitoes). The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods). The four genera of Influenza virus that infect vertebrates, which are identified by antigenic differences in their nucleoprotein and matrix protein, are as follows: * ''Alphainfluenzavirus'' infects humans, other mammals, and birds, and causes all flu pandemics * ''Betainfluenzavirus'' infects humans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Replication
Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm. Viral production / replication Viruses multiply only in living cells. The host cell must provide the energy and synthetic machinery and the low- molecular-weight precursors for the synthesis of viral proteins and nucleic acids. The virus replication occurs in seven stages, namely; # Attachment # Entry, # Uncoating, # Transcription / mRNA production, # Synthesis of virus components, # Virion assembly and # Release (Liberation Stage). Attachment It is the first step of viral replication ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PolyA
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nuclear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein. mRNA is created during the process of Transcription (biology), transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as Translation (biology), translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
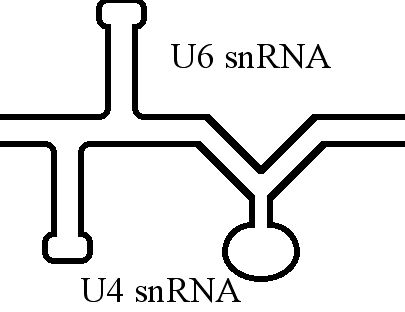
U6 SnRNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many organism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |