|
Indole Acetaldehyde
Indole-3-acetaldehyde belongs to the class of organic compounds known as indoles. These are compounds containing an indole moiety, which consists of pyrrole ring fused to benzene to form 2,3-benzopyrrole. Indole-3-acetaldehyde is a substrate for retina-specific copper amine oxidase, aldehyde dehydrogenase X (mitochondrial), amine oxidase B, amiloride-sensitive amine oxidase, aldehyde dehydrogenase (mitochondrial), fatty aldehyde dehydrogenase Fatty aldehyde dehydrogenase (or Long-chain-aldehyde dehydrogenase) is an aldehyde dehydrogenase enzyme that in human is encoded in the ALDH3A2 gene on chromosome 17. Aldehyde dehydrogenase enzymes function to remove toxic aldehydes that are gene ..., 4-trimethylaminobutyraldehyde dehydrogenase, aldehyde dehydrogenase (dimeric NADP-preferring), aldehyde dehydrogenase family 7 member A1, amine oxidase A, aldehyde dehydrogenase 1A3 and membrane copper amine oxidase. References {{reflist External links Indoleacetaldehyde (HMDB0 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Organic Compound
In chemistry, organic compounds are generally any chemical compounds that contain carbon-hydrogen or carbon-carbon bonds. Due to carbon's ability to catenate (form chains with other carbon atoms), millions of organic compounds are known. The study of the properties, reactions, and syntheses of organic compounds comprise the discipline known as organic chemistry. For historical reasons, a few classes of carbon-containing compounds (e.g., carbonate salts and cyanide salts), along with a few other exceptions (e.g., carbon dioxide, hydrogen cyanide), are not classified as organic compounds and are considered inorganic. Other than those just named, little consensus exists among chemists on precisely which carbon-containing compounds are excluded, making any rigorous definition of an organic compound elusive. Although organic compounds make up only a small percentage of Earth's crust, they are of central importance because all known life is based on organic compounds. Living t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aldehyde Dehydrogenase
Aldehyde dehydrogenases () are a group of enzymes that catalyse the oxidation of aldehydes. They convert aldehydes (R–C(=O)) to carboxylic acids (R–C(=O)). The oxygen comes from a water molecule. To date, nineteen ALDH genes have been identified within the human genome. These genes participate in a wide variety of biological processes including the detoxification of exogenously and endogenously generated aldehydes. Function Aldehyde dehydrogenase is a polymorphic enzyme responsible for the oxidation of aldehydes to carboxylic acids, which leave the liver and are metabolized by the body’s muscle and heart. There are three different classes of these enzymes in mammals: class 1 (low ''K''m, cytosolic), class 2 (low ''K''m, mitochondrial), and class 3 (high ''K''m, such as those expressed in tumors, stomach, and cornea). In all three classes, constitutive and inducible forms exist. ALDH1 and ALDH2 are the most important enzymes for aldehyde oxidation, and both a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
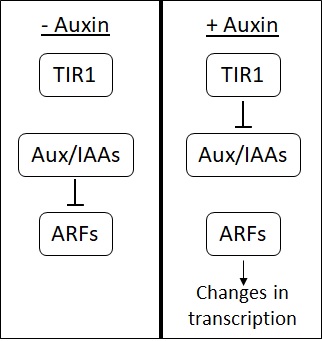
Auxins
Auxins (plural of auxin ) are a class of plant hormones (or plant-growth regulators) with some morphogen-like characteristics. Auxins play a cardinal role in coordination of many growth and behavioral processes in plant life cycles and are essential for plant body development. The Dutch biologist Frits Warmolt Went first described auxins and their role in plant growth in the 1920s. Kenneth V. Thimann became the first to isolate one of these phytohormones and to determine its chemical structure as indole-3-acetic acid (IAA). Went and Thimann co-authored a book on plant hormones, ''Phytohormones'', in 1937. Overview Auxins were the first of the major plant hormones to be discovered. They derive their name from the Greek word αυξειν (''auxein'' – "to grow/increase"). Auxin is present in all parts of a plant, although in very different concentrations. The concentration in each position is crucial developmental information, so it is subject to tight regulation through both meta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aldehyde Dehydrogenase 3 Family, Member A1
Aldehyde dehydrogenase, dimeric NADP-preferring is an enzyme that in humans is encoded by the ''ALDH3A1'' gene. Aldehyde dehydrogenases oxidize various aldehydes to the corresponding acids. They are involved in the detoxification of alcohol-derived acetaldehyde and in the metabolism of corticosteroids, biogenic amines, neurotransmitters, and lipid peroxidation. The enzyme encoded by this gene forms a cytoplasmic homodimer that preferentially oxidizes aromatic aldehyde substrates. The gene is located within the Smith–Magenis syndrome region on chromosome 17. ALDH3A1 expression is notably high in the cornea of mammalian species, comprising from 5 to 50% of soluble protein content, but is almost absent from the cornea of other vertebrates. Structure and mechanism ALDH3A1 is a homodimer consisting of alpha helices (43.8%), beta sheets (4.2%), p-loop turns (28.2%) and random coils (23.8%). The catalytic residue–Cys244—is located on an active site that contains a Rossmann fol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ALDH7A1
Aldehyde dehydrogenase 7 family, member A1, also known as ALDH7A1 or antiquitin, is an enzyme that in humans is encoded by the ''ALDH7A1'' gene. The protein encoded by this gene is a member of subfamily 7 in the aldehyde dehydrogenase gene family. These enzymes are thought to play a major role in the detoxification of aldehydes generated by alcohol metabolism and lipid peroxidation. This particular member has homology to a previously described protein from the green garden pea, the 26g pea turgor protein. It is also involved in lysine catabolism that is known to occur in the mitochondrial matrix. Recent reports show that this protein is found both in the cytosol and the mitochondria, and the two forms likely arise from the use of alternative translation initiation sites. An additional variant encoding a different isoform has also been found for this gene. Mutations in this gene are associated with pyridoxine-dependent epilepsy. Several related pseudogenes have also been identified. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aldehyde Dehydrogenase (NAD+)
In enzymology, an aldehyde dehydrogenase (NAD+) () is an enzyme that catalyzes the chemical reaction :an aldehyde + NAD+ + H2O \rightleftharpoons an acid + NADH + H+ The 3 substrates of this enzyme are aldehyde, NAD+, and H2O, whereas its 3 products are acid, NADH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the aldehyde or oxo group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is aldehyde:NAD+ oxidoreductase. Other names in common use include CoA-independent aldehyde dehydrogenase, m-methylbenzaldehyde dehydrogenase, NAD-aldehyde dehydrogenase, NAD-dependent 4-hydroxynonenal dehydrogenase, NAD-dependent aldehyde dehydrogenase, NAD-linked aldehyde dehydrogenase, propionaldehyde dehydrogenase, and aldehyde dehydrogenase (NAD). This enzyme participates in 17 metabolic pathways: glycolysis / gluconeogenesis, ascorbate and aldarate metabolism, fatty acid metabolism, bile acid biosynthesis, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
4-trimethylammoniobutyraldehyde Dehydrogenase
In enzymology, a 4-trimethylammoniobutyraldehyde dehydrogenase () is an enzyme that catalyzes the chemical reaction :4-trimethylammoniobutanal + NAD+ + H2O \rightleftharpoons 4-trimethylammoniobutanoate + NADH + 2 H+ The 3 substrates of this enzyme are 4-trimethylammoniobutanal, NAD+, and H2O, whereas its 3 products are 4-trimethylammoniobutanoate, NADH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the aldehyde or oxo group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is 4-trimethylammoniobutanal:NAD+ 1-oxidoreductase. Other names in common use include 4-trimethylaminobutyraldehyde dehydrogenase, and 4-N-trimethylaminobutyraldehyde dehydrogenase. This enzyme participates in lysine degradation and carnitine biosynthesis. See also *Carnitine biosynthesis Carnitine biosynthesis is a method for the endogenous production of L-carnitine, a molecule that is essential for energy metabolism. In huma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fatty Aldehyde Dehydrogenase
Fatty aldehyde dehydrogenase (or Long-chain-aldehyde dehydrogenase) is an aldehyde dehydrogenase enzyme that in human is encoded in the ALDH3A2 gene on chromosome 17. Aldehyde dehydrogenase enzymes function to remove toxic aldehydes that are generated by the metabolism of alcohol and by lipid peroxidation. Structure The ALDH3A2 belongs to the aldehyde dehydrogenase superfamily and is a membrane-associated protein typically containing 485 residues. The mature protein functions as a dimer. The structure was resolved using X-ray crystallography at 2.1 Angstrom resolution. It contains an element in the C-terminal region referred to as a "gatekeeper" helix, which is adjacent to the membrane-anchored transmembrane domain and the catalytic core. The gatekeeper helix appears to control access of molecular substrates to the catalytic core and allows efficient transit between membranes and catalytic sites. Function ALDH3A2 catalyzes the oxidation of long-chain aliphatic aldehydes int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amine Oxidase
An amine oxidase is an enzyme that catalyzes the oxidative cleavage of alkylamines into aldehydes and ammonia Ammonia is an inorganic compound of nitrogen and hydrogen with the formula . A stable binary hydride, and the simplest pnictogen hydride, ammonia is a colourless gas with a distinct pungent smell. Biologically, it is a common nitrogenous was ...: :RCH2NH2 + H2O + O2 \rightleftharpoons RCHO + NH3 + H2O2 Amine oxidases are divided into two subfamilies based on the cofactor they contain: References External links * {{oxidoreductase-stub EC 1.4.3 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indole
Indole is an aromatic heterocyclic organic compound with the formula C8 H7 N. It has a bicyclic structure, consisting of a six-membered benzene ring fused to a five-membered pyrrole ring. Indole is widely distributed in the natural environment and can be produced by a variety of bacteria. As an intercellular signal molecule, indole regulates various aspects of bacterial physiology, including spore formation, plasmid stability, resistance to drugs, biofilm formation, and virulence. The amino acid tryptophan is an indole derivative and the precursor of the neurotransmitter serotonin. General properties and occurrence Indole is a solid at room temperature. It occurs naturally in human feces and has an intense fecal odor. At very low concentrations, however, it has a flowery smell, and is a constituent of many perfumes. It also occurs in coal tar. The corresponding substituent is called indolyl. Indole undergoes electrophilic substitution, mainly at position 3 (see diagra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondria
A mitochondrion (; ) is an organelle found in the Cell (biology), cells of most Eukaryotes, such as animals, plants and Fungus, fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into mitosome, other structures. One eukaryote, ''Monocercomonoides'', is known to have completely lost its mitocho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ALDH1B1
Aldehyde dehydrogenase X, mitochondrial is an enzyme that in humans is encoded by the ''ALDH1B1'' gene. Function This protein belongs to the aldehyde dehydrogenases family of proteins. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. This gene does not contain introns in the coding sequence. The variation of this locus may affect the development of alcohol-related problems. Model organisms Model organisms have been used in the study of ALDH1B1 function. A conditional knockout mouse line called ''Aldh1b1tm2a(EUCOMM)Wtsi'' was generated at the Wellcome Trust Sanger Institute. Male and female animals underwent a standardized phenotypic screen In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological proper ... to determine the effects of deletion. Ad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |