|
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter Gene expression, expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Basic Units Of Chromatin Structure
BASIC (Beginners' All-purpose Symbolic Instruction Code) is a family of general-purpose, high-level programming languages designed for ease of use. The original version was created by John G. Kemeny and Thomas E. Kurtz at Dartmouth College in 1963. They wanted to enable students in non-scientific fields to use computers. At the time, nearly all computers required writing custom software, which only scientists and mathematicians tended to learn. In addition to the program language, Kemeny and Kurtz developed the Dartmouth Time Sharing System (DTSS), which allowed multiple users to edit and run BASIC programs simultaneously on remote terminals. This general model became very popular on minicomputer systems like the PDP-11 and Data General Nova in the late 1960s and early 1970s. Hewlett-Packard produced an entire computer line for this method of operation, introducing the HP2000 series in the late 1960s and continuing sales into the 1980s. Many early video games trace their h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIST1H4H
Histone H4 is a protein that in humans is encoded by the ''HIST1H4H'' gene In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba .... Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Novo Mutation
A de novo mutation is any mutation/alteration in the genome of any organism (humans, animals, plant, microbes, etc.) that wasn't present or transmitted by their parents. This type of mutation (like any other) occurs spontaneously during the process of DNA replication during cell division in a fetus whose close, biological relatives don't have the mutation. Often, these kind of mutations have very little to no effect on the affected organism, but in rare cases they have a notable and/or serious effect on overall health, physical appearance, etc. Rate The average number of spontaneous mutations (not present in the parents) an infant has in its genome is approximately 43.86 DNMs. A study done in September of 2019 by the University of Utah Health revealed that certain families have a higher spontaneous mutation rate than average, meaning that their newborns had more spontaneous mutations (not present in their parents) than the average newborn, this tendency was found to be hereditar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
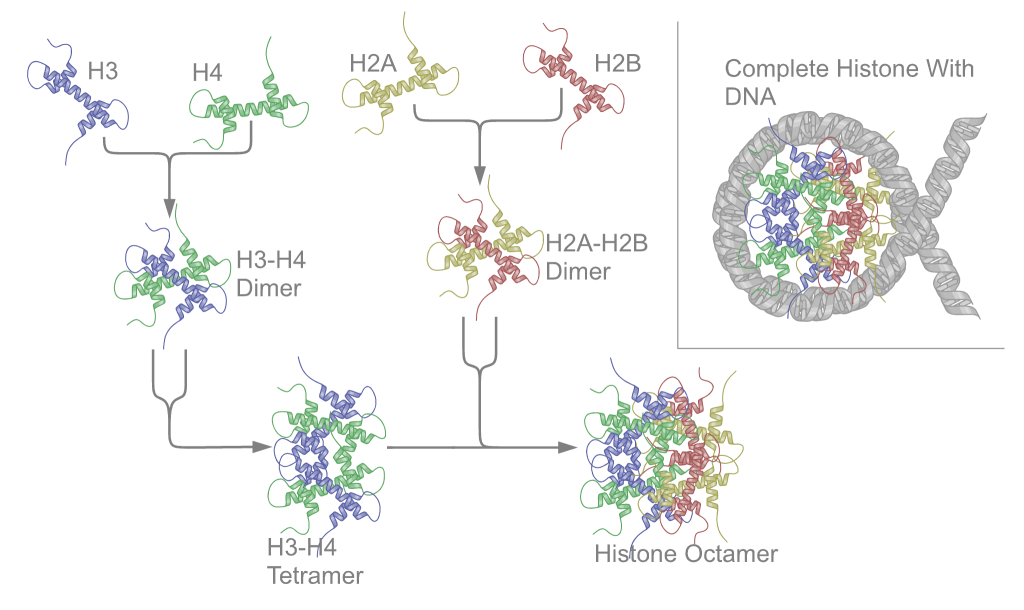
Histone Octamer
A histone octamer is the eight-protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins ( H2A, H2B, H3, and H4). The octamer assembles when a tetramer, containing two copies of H3 and two of H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Each of these key components interacts with DNA in its own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and the histone octamer loosely associated, and ultimately allow the two to re-position or to separate entirely. History of research Histone post-translational modifications were first identified and listed as having a potential regulatory role on the synthesis of RNA in 1964. Since then, over several decades, chromatin theory has evolved. Chromatin subunit models as well as the notion of the nucleosome were established in 197 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetramer
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula Ti(OCH3)4, which is tetrameric in solid state and has the molecular formula Ti4(OCH3)16. An example from organic chemistry is kobophenol A, a substance that is formed by combining four molecules of resveratrol. In biochemistry, it similarly refers to a biomolecule formed of four units, that are the same (homotetramer), i.e. as in Concanavalin A or different (heterotetramer), i.e. as in hemoglobin. Hemoglobin has 4 similar sub-units while immunoglobulins have 2 very different sub-units. The different sub-units may have each their own activity, such as binding biotin in avidin tetramers, or have a common biological property, such as the allosteric binding of oxygen in hemoglobin. See also * Cluster chemistry; atomic and molecular clusters ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalent attachment ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Fold
A histone fold is a structurally conserved motif found near the C-terminus in every core histone sequence in a histone octamer responsible for the binding of histones into heterodimers. The histone fold averages about 70 amino acids and consists of three alpha helices connected by two short, unstructured loops. When not in the presence of DNA, the core histones assemble into head-to-tail intermediates ( H3 and H4 first assemble into heterodimers then fuse two heterodimers to form a tetramer, while H2A and H2B form heterodimers) via extensive hydrophobic interactions between each histone fold domain in a "handshake motif". Also the histone fold was first found in TATA box-binding protein-associated factors, which is a main component in transcription. The histone fold’s evolution can be found by different combinations of ancestral sets of peptides that make up helix-strand-helix motif that come from the three folds from the ancestral fragments. These peptide chains can be found ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIST4H4
Histone H4 is a protein that in humans is encoded by the ''HIST4H4'' gene. Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryote Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...s. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. References Further reading * * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIST2H4A
Histone H4 is a protein that in humans is encoded by the ''HIST2H4A'' gene In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba .... Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. This structure consists of approximately 146 bp of DNA wrapped around a nucleosome, an octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in a his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIST1H4L
Histone H4 is a protein that in humans is encoded by the ''HIST1H4L'' gene. Function Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones ( H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a member of the histone H4 family. Transcripts from this gene lack polyA Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ... tails but instead contain a palindromic termination element. This gene is found in the sm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |