|
Heterochromatin Protein 1
The family of heterochromatin protein 1 (HP1) ("Chromobox Homolog", CBX) consists of highly conserved proteins, which have important functions in the cell nucleus. These functions include gene repression by heterochromatin formation, transcriptional activation, regulation of binding of cohesion complexes to centromeres, sequestration of genes to the nuclear periphery, transcriptional arrest, maintenance of heterochromatin integrity, gene repression at the single nucleosome level, gene repression by heterochromatization of euchromatin, and DNA repair. HP1 proteins are fundamental units of heterochromatin packaging that are enriched at the centromeres and telomeres of nearly all eukaryotic chromosomes with the notable exception of budding yeast, in which a yeast-specific silencing complex of SIR (silent information regulatory) proteins serve a similar function. Members of the HP1 family are characterized by an N-terminal chromodomain and a C-terminal chromoshadow domain, se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CBX5 (gene)
Chromobox protein homolog 5 is a protein that in humans is encoded by the ''CBX5'' gene. It is a highly conserved, non-histone protein part of the heterochromatin family. The protein itself is more commonly called (in humans) HP1α. Heterochromatin protein-1 (HP1) has an N-terminal domain that acts on methylated lysines residues leading to epigenetic repression. The C-terminal of this protein has a chromo shadow-domain (CSD) that is responsible for homodimerizing, as well as interacting with a variety of chromatin-associated, non-histone proteins. Structure HP1α is 191 amino acids in length containing 6 exons. As mentioned above, this protein contains two domains, an N-terminal chromodomain (CD) and a C- terminal chromoshadow domain (CSD). The CD binds with histone 3 through a methylated lysine residue at position 9 (H3K9) while the C-terminal CSD homodimerizes and interacts with a variety of other chromatin-associated, non-histone related proteins. Connecting these two domains ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sarah Elgin
Sarah C.R. Elgin is an American biochemist and geneticist. She is the Viktor Hamburger Professor of biology at Washington University in St. Louis, and is noted for her work in epigenetics, gene regulation, and heterochromatin, and for her contributions to science education. Early life and education Sarah "Sally" Elgin was born in Washington, DC. She grew up in Salem, Oregon. In high school, Elgin studied fallout levels in Oregon rainwater after nuclear weapons tests in the Soviet Union. She received her B.A. in chemistry from Pomona College in 1967. While at Pomona, she participated in a summer research program at the University of Leeds characterizing the egg stalk of the green lacewing fly Chrysopa vittata. Elgin did her graduate work in the lab of James Bonner at the California Institute of Technology, isolating and characterizing nonhistone chromosomal proteins from rat livers. She received her Ph.D. in biochemistry in 1972. Elgin stayed at Caltech for her postdoctoral r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MECP2
''MECP2'' (methyl CpG binding protein 2) is a gene that encodes the protein MECP2. MECP2 appears to be essential for the normal function of nerve cells. The protein seems to be particularly important for mature nerve cells, where it is present in high levels. The MECP2 protein is likely to be involved in turning off ("repressing" or "silencing") several other genes. This prevents the genes from making proteins when they are not needed. Recent work has shown that MECP2 can also activate other genes. The MECP2 gene is located on the long (q) arm of the X chromosome in band 28 ("Xq28"), from base pair 152,808,110 to base pair 152,878,611. MECP2 is an important reader of DNA methylation. Its methyl-CpG-binding (MBD) domain recognizes and binds 5-mC regions. MECP2 is X-linked and subject to X inactivation. MECP2 gene mutations are the cause of most cases of Rett syndrome, a progressive neurologic developmental disorder and one of the most common causes of cognitive disability in f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor. Classification Substrate MTases can be divided into three different groups on the basis of the chemical reactions they catalyze: * m6A - those that generate N6-methyladenine * m4C - those that generate N4-methylcytosine * m5C - those that generate C5-methylcytosine m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes). m5C methyltransfereases are found in some lower eukaryotes, in most higher plants, and in animals beginning with the echinoderms. The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) are enzymes that specifically methylate the amino group at the C-6 position of adenines ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, HIST2H4B, HIST4H4. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H1
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species. Structure Metazoan H1 proteins feature a central globular "winged helix" domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved. H1 found in protists and bacteria, otherwise known as nucleoproteins HC1 and HC2 (, ), lack the central domain and the N-terminal tail. H1 is less conserved than core histones. The globular domain is the most conserved part of H1. Function Unlike the other histones, H1 does not make up the nucleosome "bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. H1 is present in half the amount of the other four histones, which contribute two molecules ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalent attach ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Methyltransferase
Histone methyltransferases (HMT) are histone-modifying enzymes (e.g., histone-lysine N-methyltransferases and histone-arginine N-methyltransferases), that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific (which can be SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain containing or non-SET domain containing) and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group. The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrahymena
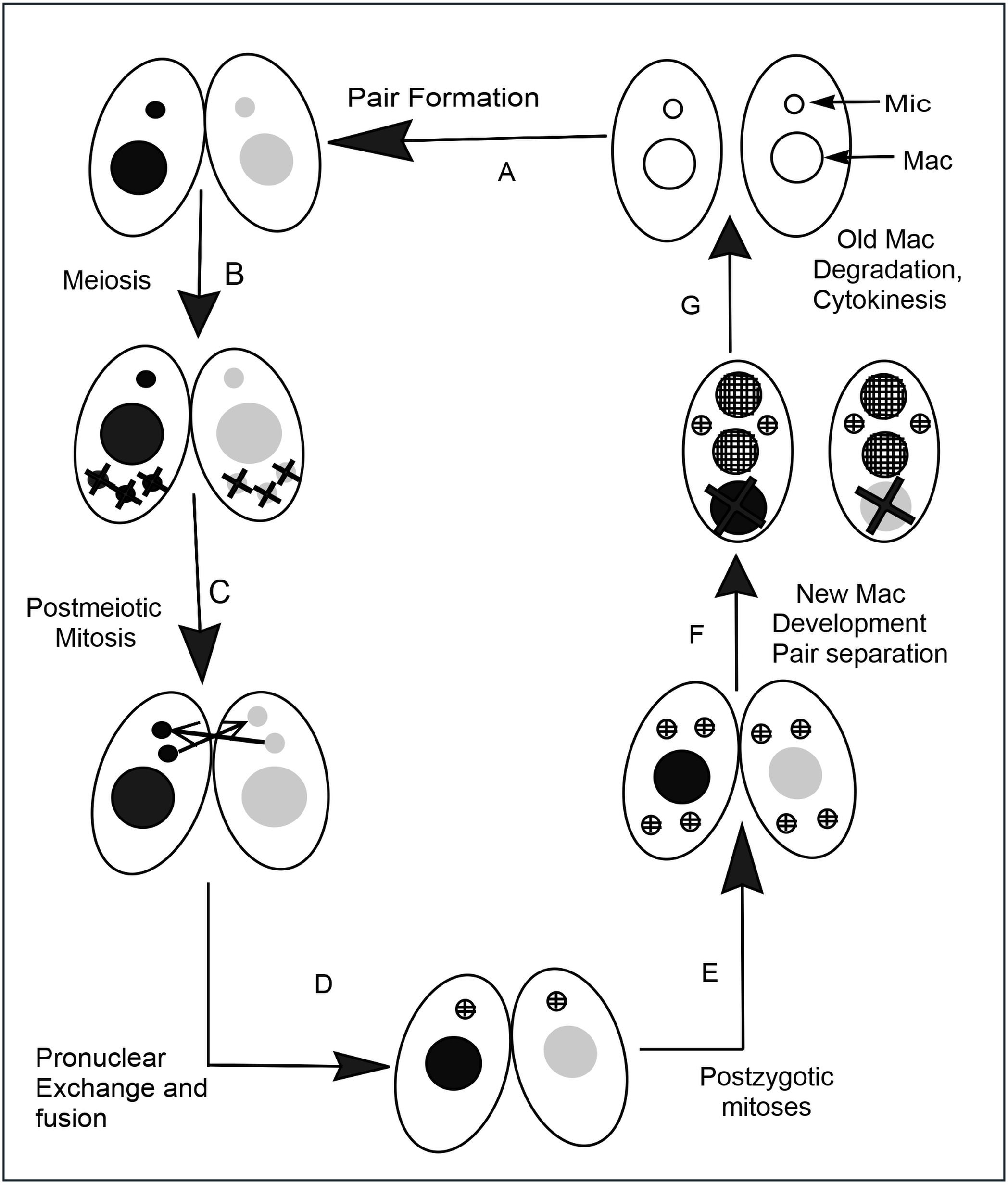
''Tetrahymena'', a unicellular eukaryote, is a genus of free-living ciliates. The genus Tetrahymena is the most widely studied member of its phylum. It can produce, store and react with different types of hormones. Tetrahymena cells can recognize both related and hostile cells. They can also switch from commensalistic to pathogenic modes of survival. They are common in freshwater lakes, ponds, and streams. ''Tetrahymena'' species used as model organisms in biomedical research are ''T. thermophila'' and '' T. pyriformis''. ''T. thermophila'': a model organism in experimental biology As a ciliated protozoan, ''Tetrahymena thermophila'' exhibits nuclear dimorphism: two types of cell nuclei. They have a bigger, non-germline macronucleus and a small, germline micronucleus in each cell at the same time and these two carry out different functions with distinct cytological and biological properties. This unique versatility allows scientists to use ''Tetrahymena'' to identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chicken
The chicken (''Gallus gallus domesticus'') is a domesticated junglefowl species, with attributes of wild species such as the grey and the Ceylon junglefowl that are originally from Southeastern Asia. Rooster or cock is a term for an adult male bird, and a younger male may be called a cockerel. A male that has been castrated is a capon. An adult female bird is called a hen and a sexually immature female is called a pullet. Humans now keep chickens primarily as a source of food (consuming both their meat and eggs) and as pets. Traditionally they were also bred for cockfighting, which is still practiced in some places. Chickens are one of the most common and widespread domestic animals, with a total population of 23.7 billion , up from more than 19 billion in 2011. There are more chickens in the world than any other bird. There are numerous cultural references to chickens – in myth, folklore and religion, and in language and literature. Genetic studies have poi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Xenopus
''Xenopus'' () (Gk., ξενος, ''xenos''=strange, πους, ''pous''=foot, commonly known as the clawed frog) is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-known species of this genus are ''Xenopus laevis'' and '' Xenopus tropicalis'', which are commonly studied as model organisms for developmental biology, cell biology, toxicology, neuroscience and for modelling human disease and birth defects. The genus is also known for its polyploidy, with some species having up to 12 sets of chromosomes. Characteristics ''Xenopus laevis'' is a rather inactive creature. It is incredibly hardy and can live up to 15 years. At times the ponds that ''Xenopus laevis'' is found in dry up, compelling it, in the dry season, to burrow into the mud, leaving a tunnel for air. It may lie dormant for up to a year. If the pond dries up in the rainy season, ''Xenopus laevis'' may migrate long distances to another pond, m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Schizosaccharomyces Pombe
''Schizosaccharomyces pombe'', also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically measure 3 to 4 micrometres in diameter and 7 to 14 micrometres in length. Its genome, which is approximately 14.1 million base pairs, is estimated to contain 4,970 protein-coding genes and at least 450 non-coding RNAs. These cells maintain their shape by growing exclusively through the cell tips and divide by medial fission to produce two daughter cells of equal size, which makes them a powerful tool in cell cycle research. Fission yeast was isolated in 1893 by Paul Lindner from East African millet beer. The species name ''pombe'' is the Swahili word for beer. It was first developed as an experimental model in the 1950s: by Urs Leupold for studying genetics, and by Murdoch Mitchison for studying the cell cycle. Paul Nurse, a fission yeast ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |