|
Hartmut Oschkinat
Hartmut Oschkinat (born 28 February 1957) is a German structural biologist and professor for chemistry at the Free University of Berlin. His research focuses on the study of biological systems with solid-state nuclear magnetic resonance. He is a member of the Editorial Boards of the ''Journal of Biomolecular NMR'' and ''Structure''. Life and career Oschkinat studied chemistry at the Johann-Wolfgang-Goethe University of Frankfurt. He received his doctoral degree in 1986 under the supervision of Horst Kessler with the title "Analysis of the conformation of Cyclosporin in solution using NMR-spectroscopy: development and use of new methods." in the field of nuclear magnetic resonance spectroscopy (NMR). After his work as a postdoctoral researcher with Geoffrey Bodenhausen at the University of Lausanne, Oschkinat moved to the Max Planck Institute of Biochemistry in Martinsried in 1987 as a post-doc in the Lab of Marius Clore and Angela Gronenborn. There, he was working with Mar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Technical University Of Munich
The Technical University of Munich (TUM or TU Munich; german: Technische Universität München) is a public research university in Munich, Germany. It specializes in engineering, technology, medicine, and applied and natural sciences. Established in 1868 by King Ludwig II of Bavaria, the university now has additional campuses in Garching, Freising, Heilbronn, Straubing, and Singapore, with the Garching campus being its largest. The university is organized into eight schools and departments, and is supported by numerous research centers. It is one of the largest universities in Germany, with 50,000 students and an annual budget of €1,770.3 million (including university hospital). A ''University of Excellence'' under the German Universities Excellence Initiative, TUM is considered the top university in Germany according to major rankings as of 2022 and is among the leading universities in the European Union. Its researchers and alumni include 18 Nobel laureates and 23 Leib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amyloid
Amyloids are aggregates of proteins characterised by a Fibril, fibrillar morphology of 7–13 Nanometer, nm in diameter, a beta sheet (β-sheet) Secondary structure of proteins, secondary structure (known as cross-β) and ability to be Staining, stained by particular dyes, such as Congo red. In the human body, amyloids have been linked to the development of various diseases. Pathogenic amyloids form when previously healthy proteins lose their normal Protein structure, structure and physiology, physiological functions (Protein misfolding, misfolding) and form fibrous deposits in amyloid plaques around cells which can disrupt the healthy function of tissues and organs. Such amyloids have been associated with (but not necessarily as the cause of) more than 50 human diseases, known as amyloidosis, and may play a role in some neurodegenerative diseases. Some of these diseases are mainly sporadic and only a few cases are Genetic disorder, familial. Others are only Genetic disorder, fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
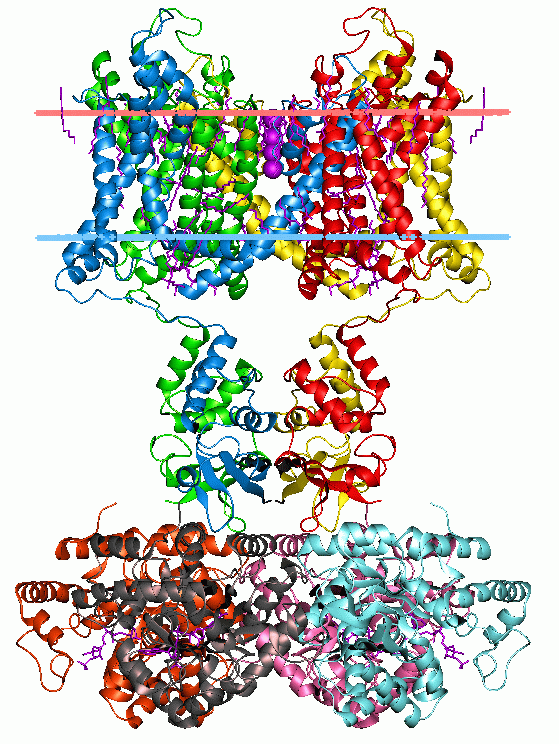
Membrane Proteins
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the surv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SH3 Domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Magic Angle Spinning
In solid-state NMR spectroscopy, magic-angle spinning (MAS) is a technique routinely used to produce better resolution NMR spectra. MAS NMR consists in spinning the sample (usually at a frequency of 1 to 130 kHz) at the magic angle θm (ca. 54.74°, where cos2θm=1/3) with respect to the direction of the magnetic field. Three main interactions responsible in solid state NMR (dipolar, chemical shift anisotropy, quadrupolar) often lead to very broad and featureless NMR lines. However, these three interactions in solids are orientation-dependent and can be averaged to some extent by MAS: * The nuclear dipolar interaction has a 3\cos^2\theta - 1 dependence, where \theta is the angle between the internuclear axis and the main magnetic field. As a result, the dipolar interaction vanish at the magic angle θm and the interaction contributing to the line broadening is removed. Even though all internuclear vectors cannot be all set to the magic angle, rotating the sample around this ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solid-state NMR
Solid-state NMR (ssNMR) spectroscopy is a technique for characterizing atomic level structure in solid materials e.g. powders, single crystals and amorphous samples and tissues using nuclear magnetic resonance (NMR) spectroscopy. The anisotropic part of many spin interactions are present in solid-state NMR, unlike in solution-state NMR where rapid tumbling motion averages out many of the spin interactions. As a result, solid-state NMR spectra are characterised by larger linewidths than in solution state NMR, which can be utilized to give quantitative information on the molecular structure, conformation and dynamics of the material. Solid-state NMR is often combined with magic angle spinning to remove anisotropic interactions and improve the resolution as well as the sensitivity of the technique. Nuclear spin interactions The resonance frequency of a nuclear spin depends on the strength of the magnetic field at the nucleus, which can be modified by isotropic (e.g. chemical shift, is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fragment-based Drug Discovery
Fragment-based lead discovery (FBLD) also known as fragment-based drug discovery (FBDD) is a method used for finding lead compounds as part of the drug discovery process. Fragments are small organic molecules which are small in size and low in molecular weight. It is based on identifying small chemical fragments, which may bind only weakly to the biological target, and then growing them or combining them to produce a lead with a higher affinity. FBLD can be compared with high-throughput screening (HTS). In HTS, libraries with up to millions of compounds, with molecular weights of around 500 atomic mass unit, Da, are screened, and nanomolar binding affinities are sought. In contrast, in the early phase of FBLD, libraries with a few thousand compounds with molecular weights of around 200 Da may be screened, and millimolar affinities can be considered useful. FBLD is a technique being used in research for discovering novel potent Enzyme inhibitor, inhibitors. This methodology could help ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WW Domain
The WW domain, (also known as the rsp5-domain or WWP repeating motif) is a modular protein domain that mediates specific interactions with protein ligands. This domain is found in a number of unrelated signaling and structural proteins and may be repeated up to four times in some proteins. Apart from binding preferentially to proteins that are proline-rich, with particular proline-motifs, PP-P- PY, some WW domains bind to phosphoserine- phosphothreonine-containing motifs. Structure and ligands The WW domain is one of the smallest protein modules, composed of only 40 amino acids, which mediates specific protein-protein interactions with short proline-rich or proline-containing motifs. Named after the presence of two conserved tryptophans (W), which are spaced 20-22 amino acids apart within the sequence, the WW domain folds into a meandering triple-stranded beta sheet. The identification of the WW domain was facilitated by the analysis of two splice isoforms of YAP gene prod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pleckstrin Homology Domain
Pleckstrin homology domain (PH domain) or (PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton. This domain can bind phosphatidylinositol lipids within biological membranes (such as phosphatidylinositol (3,4,5)-trisphosphate and phosphatidylinositol (4,5)-bisphosphate), and proteins such as the βγ-subunits of heterotrimeric G proteins, and protein kinase C. Through these interactions, PH domains play a role in recruiting proteins to different membranes, thus targeting them to appropriate cellular compartments or enabling them to interact with other components of the signal transduction pathways. Lipid binding specificity Individual PH domains possess specificities for phosphoinositides phosphorylated at different sites within the inositol ring, e.g., some bind phosphatidylinositol (4,5)-bisphosphate but not phosphatidylinositol (3,4,5)-trisphosphate or p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |