|
HIV Ribosomal Frameshift Signal
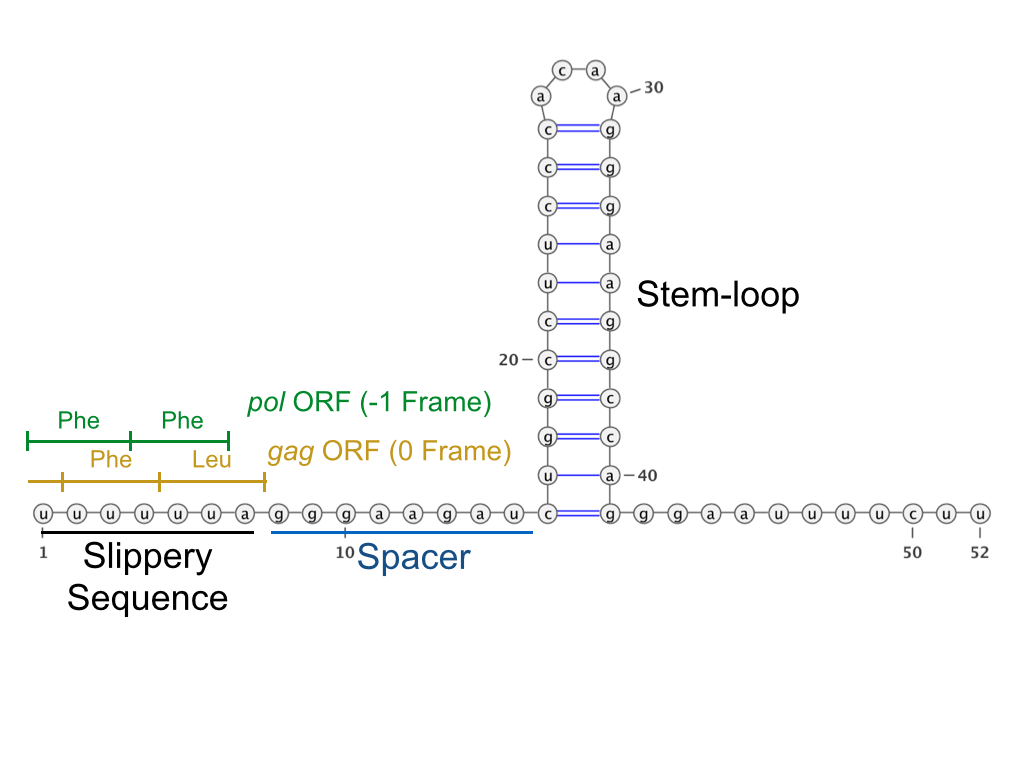
HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence. Intact and consistent protein biosynthesis relies on the ability of the ribosome to stay in the correct open reading frame (ORF) during translation. When the ribosome fails to maintain the proper ORF, translation usually results in either incorrect protein synthesis or early termination as a result of the introduction of a premature stop codon. However, a shift in the ORF is not universally deleterious, as many viruses capitalize on this phenomenon by using a programmed ribosomal frameshift (PRF) to translate several proteins from the same sequence, thereby maximizing the storage capacity of their genome. Thus, many viruses (including HIV-1) are categorized as having a polycistronic genome, meaning they employ multiple active ORF's in a single gene. The HIV-1 virus requires a programmed -1 ribosomal frame ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-acting Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ty ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream And Downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to relative positions of genetic code in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbon position on the deoxyribose (or ribose) ring. By convention, upstream and downstream relate to the 5' to 3' direction respectively in which RNA transcription takes place. Upstream is toward the 5' end of the RNA molecule and downstream is toward the 3' end. When considering double-stranded DNA, upstream is toward the 5' end of the coding strand for the gene in question and downstream is toward the 3' end. Due to the anti-parallel nature of DNA, this means the 3' end of the template strand is upstream of the gene and the 5' end is downstream. Some genes on the same DNA molecule may be transcribed in opposite directions. This means the upstream and downstream areas of the molecule may change depending on which gene is used as the reference. See also *Upstream and downstream (transdu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Slippery Sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame. A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins. The frameshift occurs due to wobble pairing. The Gibbs free energy of secondary structures downstream give a hint at how often frameshift happens. Tension on the mRNA molecule also plays a role. A list of slippery sequences found in animal viruses is available from Huang et al. Slippery sequences that cause a 2-base slip (−2 frameshift) have been constructed out of the HIV UUUUUUA sequence. See also *Nuclei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Structure
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary. Primary structure Primary structure consists of a linear sequence of nucleotides that are linked together by phosphodiester bond. It is this linear sequence of nucleotides that make up the primary structure of DNA or RNA. Nucleotides consist of 3 components: # Nitrogenous base ## Adenine ## Guanine ## Cytosine ## Thymine (present in DNA only) ## Uracil (present in RNA only) # 5-carbon sugar which is called deoxyribose (found in DNA) and ribose (found in RNA). # One or more phosphate groups. The nitrogen bases adenine and guanine are purine in structure and form a glycosidic bond between their 9 nitrogen and the 1' -OH group of the deoxyribose. Cytosine, thymine, and uracil are pyrimidines, hence the glycosidic bonds form betwee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiviral Drug
Antiviral drugs are a class of medication used for treating viral infections. Most antivirals target specific viruses, while a broad-spectrum antiviral is effective against a wide range of viruses. Unlike most antibiotics, antiviral drugs do not destroy their target pathogen; instead they inhibit its development. Antiviral drugs are one class of antimicrobials, a larger group which also includes antibiotic (also termed antibacterial), antifungal and antiparasitic drugs, or antiviral drugs based on monoclonal antibodies. Most antivirals are considered relatively harmless to the host, and therefore can be used to treat infections. They should be distinguished from viricides, which are not medication but deactivate or destroy virus particles, either inside or outside the body. Natural viricides are produced by some plants such as eucalyptus and Australian tea trees. Medical uses Most of the antiviral drugs now available are designed to help deal with HIV, herpes viruses, th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integrase
Retroviral integrase (IN) is an enzyme produced by a retrovirus (such as HIV) that integrates—forms covalent links between—its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases (recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination. The macromolecular complex of an IN macromolecule bound to the ends of the viral DNA ends has been referred to as the '' intasome''; IN is a key component in this and the retroviral pre-integration complex. Structure All retroviral IN proteins contain three canonical domains, connected by flexible linkers: ● an N-terminal HH-CC zinc-binding domain (a three-helical bundle stabilized by coordination of a Zn(II) cation) ● a catalytic core domain (RNaseH fold) ● a C-terminal DNA-binding domain ( SH3 fold). Crystal and NMR structures of the individual domains and 2-domain constructs of integrases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIV-1 Protease
HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins (namely, Gag and Gag- Pol) at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious. Structure Mature HIV protease exists as a 22 kDa homodimer, with each subunit made up of 99 amino acids. A single active site lies between the identical subunits and has the characteristic Asp- Thr- Gly (Asp25, Thr26 and Gly27) catalytic triad sequence common to aspartic proteases. As HIV-1 PR can only function as a dimer, the mature protease contains two Asp25 amino acids, one from each monomer, that act in conjunction with each other as the catalytic residues. Additionally, HIV protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)



