|
Histone Modifications
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30- nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around hist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
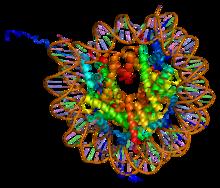
Nucleosome Structure
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electrostatic
Electrostatics is a branch of physics that studies electric charges at rest (static electricity). Since classical times, it has been known that some materials, such as amber, attract lightweight particles after rubbing. The Greek word for amber, (), was thus the source of the word 'electricity'. Electrostatic phenomena arise from the forces that electric charges exert on each other. Such forces are described by Coulomb's law. Even though electrostatically induced forces seem to be rather weak, some electrostatic forces are relatively large. The force between an electron and a proton, which together make up a hydrogen atom, is about 36 orders of magnitude stronger than the gravitational force acting between them. There are many examples of electrostatic phenomena, from those as simple as the attraction of plastic wrap to one's hand after it is removed from a package, to the apparently spontaneous explosion of grain silos, the damage of electronic components during manufacturi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pairs
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solenoid (DNA)
The solenoid structure of chromatin is a model for the structure of the 30 nm fibre. It is a secondary chromatin structure which helps to package eukaryotic DNA into the nucleus. Background Chromatin was first discovered by Walther Flemming by using aniline dyes to stain it. In 1974, it was first proposed by Roger Kornberg that chromatin was based on a repeating unit of a histone octamer and around 200 base pairs of DNA. The solenoid model was first proposed by John Finch and Aaron Klug in 1976. They used electron microscopy images and X-ray diffraction patterns to determine their model of the structure. This was the first model to be proposed for the structure of the 30 nm fibre. Structure DNA in the nucleus is wrapped around nucleosomes, which are histone octamers formed of core histone proteins; two histone H2A- H2B dimers, two histone H3 proteins, and two histone H4 proteins. The primary chromatin structure, the least-packed form, is the 11 nm, or “beads o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter Gene expression, expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalent attachment ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes. Structure Histone H2B is a lightweight structural protein made of 126 amino acids. Many of these amino acids have a positive charge at cellular pH, which allows them to interact with the negatively charged phosphate groups in DNA. Along with a central globular domain, histone H2B has two flexible histone tails that extend outwards – one at the N-terminal end and one at C-terminal end. These are highly involved in condensing chromatin from the beads-on-a-string conformation to a 30-nm fiber. Similar to other histone proteins, histone H2B has a distinct histone fold that is optimized for histone-histone as well as histone-DNA interactions. Two copies of histone H2B come together with two copies each of histone H2A, histone H3, and histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2A
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. The other histone proteins are: H1, H2B, H3 and H4. Background Histones are proteins that package DNA into nucleosomes. Histones are responsible for maintaining the shape and structure of a nucleosome. One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA. There are five families of histones known to date; these histones are termed H1/H5, H2A, H2B, H3, and H4. H2A is considered a core histone, along with H2B, H3 and H4. Core formation first occurs through the interaction of two H2A molecules. Then, H2A forms a dimer with H2B; the core molecule is complete when H3-H4 also attaches to form a tetramer. Sequence variants Histone H2A is composed of non-allelic variants. The term "Histone H2A" is intentionally non-specific and refers to a variety of closely related proteins that vary often by only a few amino acids. Apart ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H1
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species. Structure Metazoan H1 proteins feature a central globular "winged helix" domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved. H1 found in protists and bacteria, otherwise known as nucleoproteins HC1 and HC2 (, ), lack the central domain and the N-terminal tail. H1 is less conserved than core histones. The globular domain is the most conserved part of H1. Function Unlike the other histones, H1 does not make up the nucleosome "bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. H1 is present in half the amount of the other four histones, which contribute two molecules ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |