|
HECT Domain
In molecular biology, the HECT domain is a protein domain found in ubiquitin-protein ligases. The name HECT comes from 'Homologous to the E6-AP Carboxyl Terminus'. Proteins containing this domain at the C terminus include ubiquitin-protein ligase, which regulates ubiquitination of CDC25. Ubiquitin-protein ligase accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester, and then directly transfers the ubiquitin to targeted substrates. A cysteine residue is required for ubiquitin-thiolester formation. Human thyroid receptor interacting protein 12 (TRIP12), which also contains this domain, is a component of an ATP-dependent multisubunit protein that interacts with the ligand binding domain of the thyroid hormone receptor. It could be an E3 ubiquitin-protein ligase. Human E6AP ubiquitin-protein ligase interacts with the E6 protein of the cancer-associated Human papillomavirus type 16 and Human papillomavirus type 18. The E6/E6-AP complex binds to and tar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRIP12
Probable E3 ubiquitin-protein ligase TRIP12 is an enzyme that in humans is encoded by the ''TRIP12'' gene. Interactions TRIP12 has been shown to interact with APPBP1 NEDD8-activating enzyme E1 regulatory subunit is a protein that in humans is encoded by the ''NAE1'' gene. Function The protein encoded by this gene binds to the beta-amyloid precursor protein. Beta-amyloid precursor protein is a cell surface .... References Further reading * * * * * * * {{gene-2-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumour Suppressor Gene
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly landscaper genes regulate growth by contributing to the surrounding environment, when mutated can cause an envir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Binding
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding. In some cases, the associations can be quite strong—for example, the protein streptavidin and the vitamin biotin have a dissociation constant (reflecting the ratio between bound and free biotin) on the order of 10−14—and so the reactions are effectively irreversible. The result of molecular binding is sometimes the formation of a molecular complex in which the attractive forces holding the components together are generally non-covalent, and thus are normally energetically weaker than covalent bonds. Molecular binding occurs in biological complexes (e.g., between pairs or sets of proteins, or between a protein and a small molecule ligand it binds) and also in abiologic chemical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
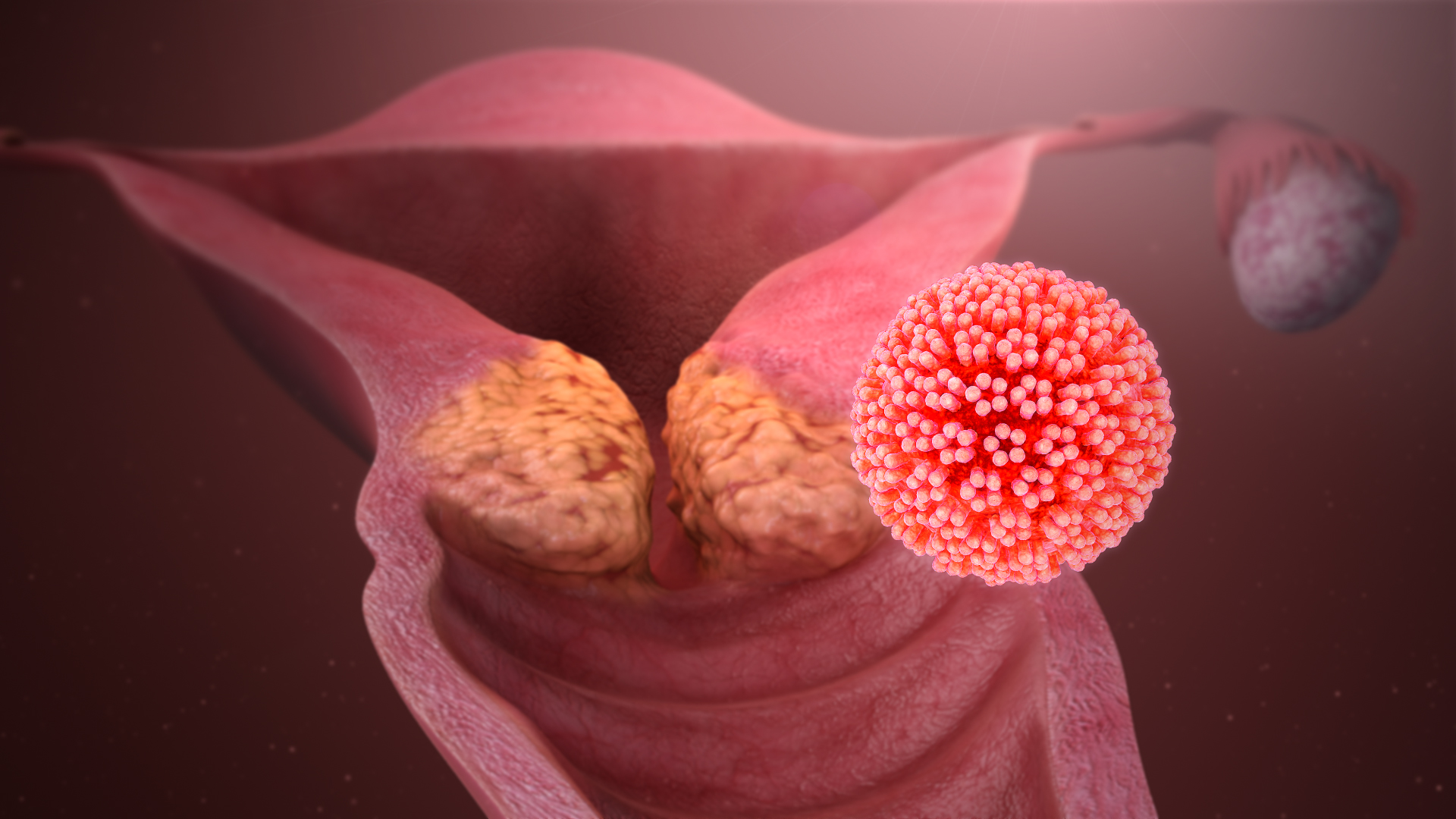
Human Papillomavirus
Human papillomavirus infection (HPV infection) is caused by a DNA virus from the ''Papillomaviridae'' family. Many HPV infections cause no symptoms and 90% resolve spontaneously within two years. In some cases, an HPV infection persists and results in either warts or precancerous lesions. These lesions, depending on the site affected, increase the risk of cancer of the cervix, vulva, vagina, penis, anus, mouth, tonsils, or throat. Nearly all cervical cancer is due to HPV and two strains – HPV16 and HPV18 – account for 70% of cases. HPV16 is responsible for almost 90% of HPV-positive oropharyngeal cancers. Between 60% and 90% of the other cancers listed above are also linked to HPV. HPV6 and HPV11 are common causes of genital warts and laryngeal papillomatosis. An HPV infection is caused by ''human papillomavirus'', a DNA virus from the papillomavirus family. Over 170 types have been described. An individual can become infected with more than one type of HPV, and the dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E6AP
Ubiquitin-protein ligase E3A (UBE3A) also known as E6AP ubiquitin-protein ligase (E6AP) is an enzyme that in humans is encoded by the ''UBE3A'' gene. This enzyme is involved in targeting proteins for degradation within cells. Protein degradation is a normal process that removes damaged or unnecessary proteins and helps maintain the normal functions of cells. Ubiquitin protein ligase E3A attaches a small marker protein called ubiquitin to proteins that should be degraded. Cellular structures called proteasomes recognize and digest proteins tagged with ubiquitin. Both copies of the UBE3A gene are active in most of the body's tissues. In most neurons, however, only the copy inherited from a person's mother (the maternal copy) is normally active; this is known as paternal imprinting. Recent evidence shows that at least some glial cells and neurons may exhibit biallelic expression of UBE3A. Further work is thus needed to delineate a complete map of UBE3A imprinting in humans and model ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thyroid Hormone Receptor
The thyroid hormone receptor (TR) is a type of nuclear receptor that is activated by binding thyroid hormone. TRs act as transcription factors, ultimately affecting the regulation of gene transcription and translation. These receptors also have non-genomic effects that lead to second messenger activation, and corresponding cellular response. Structure There are four domains that are present in all TRs. Two of these, the DNA-binding (DBD) and hinge domains, are involved in the ability of the receptor to bind hormone response elements( HREs). TRs also have a ligand binding domain (LBD) that allows them to bind to thyroid hormone with high affinity. The fourth domain is a transactivation domain which allows the receptor to bind other transcription factors. Function Thyroid hormone receptors play critical roles in the regulation of metabolism, heart rate, and development of organisms. These receptors are typically associated with retinoic acid receptors (RXR), forming heterod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand Binding
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of ligand ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar (ribose), which i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |