|
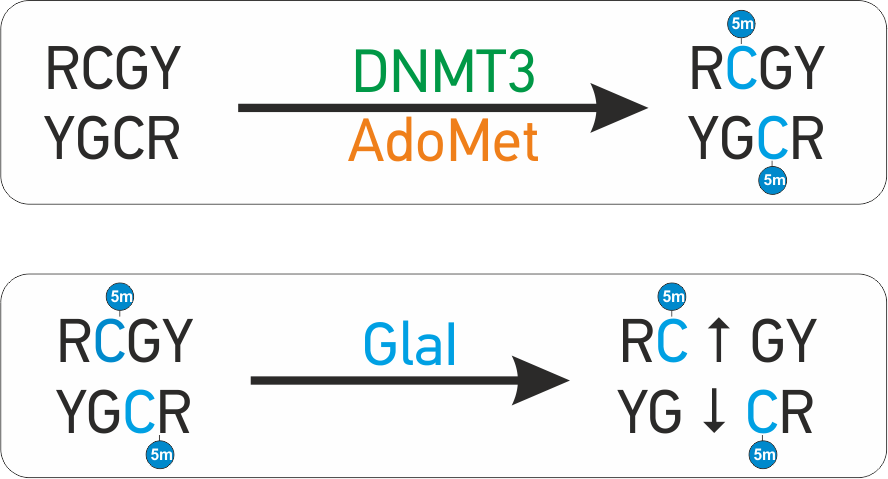
GLAD-PCR-assay
Glal hydrolysis and Ligation Adapter Dependent PCR assay (GLAD-PCR assay) is the novel method to determine R(5mC)GY sites produced in the course of ''de novo'' DNA methylation with DNMTЗA and DNMTЗB DNA methyltransferases. GLAD-PCR assay do not require bisulfite treatment of the DNA. Method was specially designed to determine methylation of RCGY site of interest in human and mammalian genomes in excess of corresponding unmethylated sites. This is a typical situation for DNA preparations from clinical samples of blood and tissues. GLAD-PCR assay is based on the new type of enzymes - site-specific methyl-directed DNA-endonucleases (MD DNA endonucleases). These enzymes are very similar to restriction enzymes in biochemical properties and cleave DNA completely, but act in opposite way: they cleave only methylated DNA and do not cleave unmethylated DNA at all. Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation. * DNMT1 maintains DNA methyla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis. As of 2016, two nucleobases have been found on which natural, enzymatic DNA methylation takes place: adenine and cytosine. The modified bases are N6-methyladenineD. B. Dunn, J. D. Smith: ''The occurrence of 6-methylaminopurine in deoxyribonucleic acids.'' In: ''Biochem J.'' 68(4), Apr 1958, S. 627–636. PMID 13522672. ., 5-methylcytosineB. F. Vanyushin, S. G. Tkacheva, A. N. Belozersky: ''Rare bases in animal DNA.'' In: ''Nature.'' 225, 1970, S. 948–949. PMID 4391887. and N4- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GLAD-PCR-assay
Glal hydrolysis and Ligation Adapter Dependent PCR assay (GLAD-PCR assay) is the novel method to determine R(5mC)GY sites produced in the course of ''de novo'' DNA methylation with DNMTЗA and DNMTЗB DNA methyltransferases. GLAD-PCR assay do not require bisulfite treatment of the DNA. Method was specially designed to determine methylation of RCGY site of interest in human and mammalian genomes in excess of corresponding unmethylated sites. This is a typical situation for DNA preparations from clinical samples of blood and tissues. GLAD-PCR assay is based on the new type of enzymes - site-specific methyl-directed DNA-endonucleases (MD DNA endonucleases). These enzymes are very similar to restriction enzymes in biochemical properties and cleave DNA completely, but act in opposite way: they cleave only methylated DNA and do not cleave unmethylated DNA at all. Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation. * DNMT1 maintains DNA methyla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor. Classification Substrate MTases can be divided into three different groups on the basis of the chemical reactions they catalyze: * m6A - those that generate N6-methyladenine * m4C - those that generate N4-methylcytosine * m5C - those that generate C5-methylcytosine m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes). m5C methyltransfereases are found in some lower eukaryotes, in most higher plants, and in animals beginning with the echinoderms. The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) are enzymes that specifically methylate the amino group at the C-6 position of adenine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzymes
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MD DNA Endonucleases
MD, Md, mD or md may refer to: Places * Moldova (ISO country code MD) * Maryland (US postal abbreviation MD) * Magdeburg (vehicle plate prefix MD), a city in Germany * Mödling District (vehicle plate prefix MD), in Lower Austria, Austria People * Muhammad (name) or Mohammed (Md) Arts, entertainment, and media Music * ' or ' (MD or m.d.; "right hand"), in piano scores * Music director * Mini Disc Other arts, entertainment, and media * ''MDs'' (TV series), 2002 * ', ("Materials and discussions for the analysis of classical texts"), an Italian journal Brands and enterprises * Air Madagascar, IATA airline code * McDonnell Douglas aircraft model prefix * MD Helicopters Science and technology Biology and medicine * Doctor of Medicine, a medical degree * Medial dorsal nucleus, a cluster of neurons in the thalamus * Muscular dystrophy, a group of diseases involving breakdown of skeletal muscles * Ménière's disease, a disorder of the inner ear * MD (Ayurveda), a degree in Indian me ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypermethylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences. In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics. Methanogenesis Methanogenesis, the process tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cpg-islands
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands (or CG islands). Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Methylated cytosines often mutate to thymines. In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a CpG island. CpG characteristics Definition ''CpG'' is shorthand for ''5'—C—phosphate—G—3' '', that is, cytosine and guanine separated by only one phosphate group; phosphate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |