GLAD-PCR-assay on:
[Wikipedia]
[Google]
[Amazon]
Glal hydrolysis and Ligation Adapter Dependent PCR assay (GLAD-PCR assay) is the novel method to determine R(5mC)GY sites produced in the course of ''de novo''  Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation.
* DNMT1 maintains DNA methylation pattern in vivo modifying a new strand after replication.
* DNMT3a and DNMT3b are responsible for DNA methylation ''de novo'' including abnormal hypermethylation in cancer cells.
It is well known that
Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation.
* DNMT1 maintains DNA methylation pattern in vivo modifying a new strand after replication.
* DNMT3a and DNMT3b are responsible for DNA methylation ''de novo'' including abnormal hypermethylation in cancer cells.
It is well known that 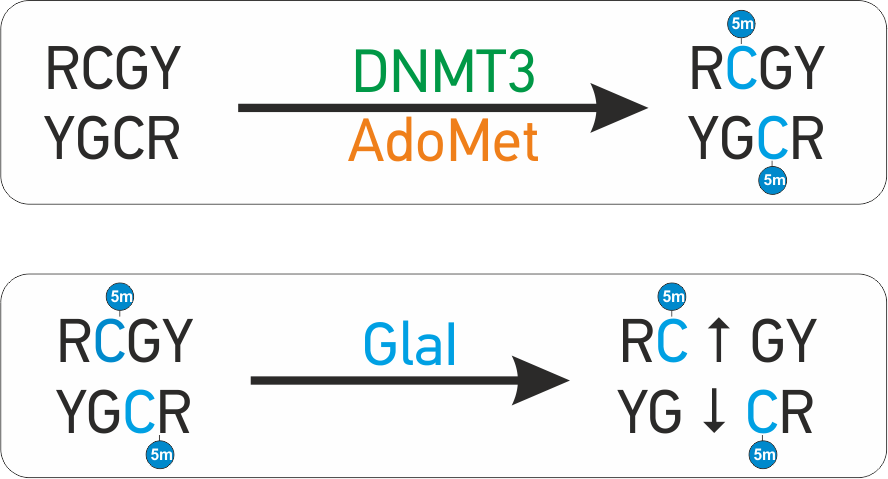 GLAD-PCR assay includes 3 simple steps:
# GlaI hydrolysis of the studied DNA. At this step only R(5mC)GY sites are hydrolyzed. Unmethylated RCGY sites remain uncut.
# The universal adapter ligation. As an adapter an oligonucleotide duplex 5’-CCTGCTCTTTCATCG-3’/3’-pGGACGAGAAAGTAGCp-5’ is used, where “p” means phosphate.
#Subsequent real-time PCR with Taqman probe. Genome primer and TaqMan probe are designed for DNA region of interest, another hybrid primer consist of two parts: one part is complementary to the universal adapter and another one - to the DNA at the point of GlaI hydrolysis.
GLAD-PCR assay includes 3 simple steps:
# GlaI hydrolysis of the studied DNA. At this step only R(5mC)GY sites are hydrolyzed. Unmethylated RCGY sites remain uncut.
# The universal adapter ligation. As an adapter an oligonucleotide duplex 5’-CCTGCTCTTTCATCG-3’/3’-pGGACGAGAAAGTAGCp-5’ is used, where “p” means phosphate.
#Subsequent real-time PCR with Taqman probe. Genome primer and TaqMan probe are designed for DNA region of interest, another hybrid primer consist of two parts: one part is complementary to the universal adapter and another one - to the DNA at the point of GlaI hydrolysis. Assay is performed in one tube, takes about 2–3 hours and determines even several copies of DNA with R(5mC)GY site of interest.
Assay is performed in one tube, takes about 2–3 hours and determines even several copies of DNA with R(5mC)GY site of interest.
DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
with DNMTЗA and DNMTЗB DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl m ...
s. GLAD-PCR assay do not require bisulfite treatment of the DNA.
Method was specially designed to determine methylation of RCGY site of interest in human and mammalian genomes in excess of corresponding unmethylated sites. This is a typical situation for DNA preparations from clinical samples of blood and tissues.
GLAD-PCR assay is based on the new type of enzymes - site-specific methyl-directed DNA-endonucleases (MD DNA endonucleases). These enzymes are very similar to restriction enzymes
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
in biochemical properties and cleave DNA completely, but act in opposite way: they cleave only methylated DNA and do not cleave unmethylated DNA at all.  Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation.
* DNMT1 maintains DNA methylation pattern in vivo modifying a new strand after replication.
* DNMT3a and DNMT3b are responsible for DNA methylation ''de novo'' including abnormal hypermethylation in cancer cells.
It is well known that
Mammalian DNA-methyltransferases DNMT1, DNMT3a and DNMT3b catalyze a reaction of DNA methylation.
* DNMT1 maintains DNA methylation pattern in vivo modifying a new strand after replication.
* DNMT3a and DNMT3b are responsible for DNA methylation ''de novo'' including abnormal hypermethylation in cancer cells.
It is well known that hypermethylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
of CpG-islands in regulatory regions of promoter and/or first exon in a variety of genes often occurs at early stages of sporadic carcinogenesis
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abnor ...
. This leads to downregulation of the genes expression in tumor cells, whereas in a healthy tissue the corresponding genes remain to be active. Thus, the detection of such epigenetic biomarkers is one of the most promising diagnostic and prognostic tools
Study of DNMT3a and DNMT3b substrate specificity has shown that both enzymes predominantly recognize RCGY site and modify internal CG-dinucleotide to form 5’-R(5mC)GY-3’/3’-YG(5mC) R-5’ sequence. One of new enzymes GlaI recognizes and cleaves site R(5mC)GY. Due to this unique substrate specificity, GlaI is a convenient tool for identification of ''de novo'' methylated sites in the human and mammalian DNA. GLAD-PCR assay includes 3 simple steps:
# GlaI hydrolysis of the studied DNA. At this step only R(5mC)GY sites are hydrolyzed. Unmethylated RCGY sites remain uncut.
# The universal adapter ligation. As an adapter an oligonucleotide duplex 5’-CCTGCTCTTTCATCG-3’/3’-pGGACGAGAAAGTAGCp-5’ is used, where “p” means phosphate.
#Subsequent real-time PCR with Taqman probe. Genome primer and TaqMan probe are designed for DNA region of interest, another hybrid primer consist of two parts: one part is complementary to the universal adapter and another one - to the DNA at the point of GlaI hydrolysis.
GLAD-PCR assay includes 3 simple steps:
# GlaI hydrolysis of the studied DNA. At this step only R(5mC)GY sites are hydrolyzed. Unmethylated RCGY sites remain uncut.
# The universal adapter ligation. As an adapter an oligonucleotide duplex 5’-CCTGCTCTTTCATCG-3’/3’-pGGACGAGAAAGTAGCp-5’ is used, where “p” means phosphate.
#Subsequent real-time PCR with Taqman probe. Genome primer and TaqMan probe are designed for DNA region of interest, another hybrid primer consist of two parts: one part is complementary to the universal adapter and another one - to the DNA at the point of GlaI hydrolysis. Assay is performed in one tube, takes about 2–3 hours and determines even several copies of DNA with R(5mC)GY site of interest.
Assay is performed in one tube, takes about 2–3 hours and determines even several copies of DNA with R(5mC)GY site of interest.
References
{{Reflist, 30em DNA Epigenetics Methylation Molecular biology