|
Gal4-UAS
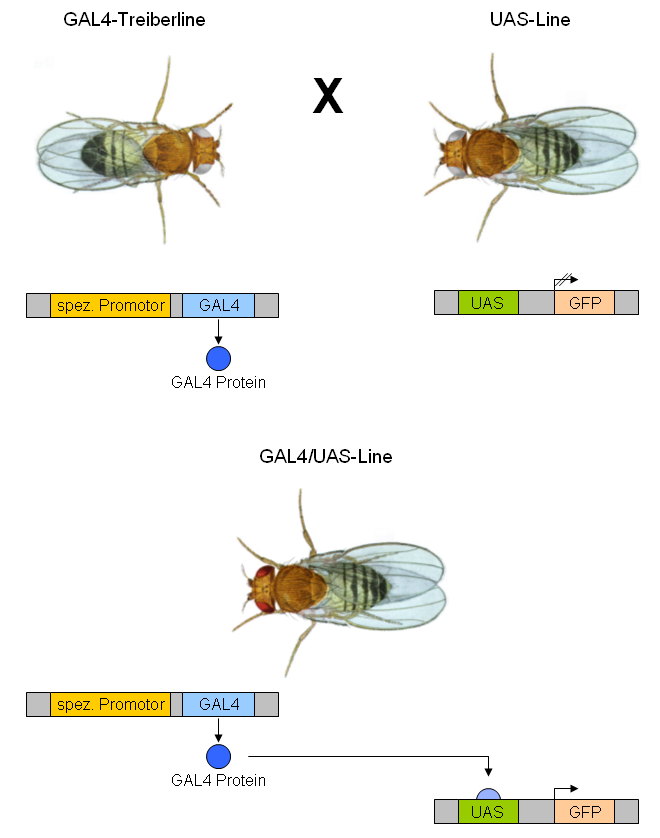
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS ( Upstream Activation Sequence), an enhancer to which GAL4 specifically binds to activate gene transcription. Overview The Gal4 system allows separation of the problems of defining which cells express a gene or protein and what the experimenter wants to do with this knowledge. Geneticists have created genetic variants of model organisms (typically fruit flies), called ''GAL4 lines'', each of which expresses GAL4 i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pigment Dispersing Factor
''Pigment dispersing factor'' (''pdf'') is a gene that encodes the protein PDF, which is part of a large family of neuropeptides. Its hormonal product, pigment dispersing hormone (PDH), was named for the diurnal pigment movement effect it has in crustacean retinal cells upon its initial discovery in the central nervous system of arthropods. The movement and aggregation of pigments in retina cells and extra-retinal cells is hypothesized to be under a split hormonal control mechanism. One hormonal set is responsible for concentrating chromatophoral pigment by responding to changes in the organism's exposure time to darkness. Another hormonal set is responsible for dispersion and responds to the light cycle. However, insect ''pdf'' genes do not function in such pigment migration since they lack the chromatophore.The Interactive Fl2011 Apr 28. The gene was first isolated and studied in '' Drosophila melanogaster, Drosophila'' by Jeffrey C. Hall's laboratory at Brandeis University ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Channelrhodopsin
Channelrhodopsins are a subfamily of retinylidene proteins ( rhodopsins) that function as light-gated ion channels. They serve as sensory photoreceptors in unicellular green algae, controlling phototaxis: movement in response to light. Expressed in cells of other organisms, they enable light to control electrical excitability, intracellular acidity, calcium influx, and other cellular processes (see optogenetics). Channelrhodopsin-1 (ChR1) and Channelrhodopsin-2 (ChR2) from the model organism ''Chlamydomonas reinhardtii'' are the first discovered channelrhodopsins. Variants that are sensitive to different colors of light or selective for specific ions (ACRs, KCRs) have been cloned from other species of algae and protists. History Phototaxis and photoorientation of microalgae have been studied over more than hundred years in many laboratories worldwide. In 1980, Ken Foster developed the first consistent theory about the functionality of algal eyes. He also analyzed published ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circadian Rhythm
A circadian rhythm (), or circadian cycle, is a natural, internal process that regulates the sleep–wake cycle and repeats roughly every 24 hours. It can refer to any process that originates within an organism (i.e., Endogeny (biology), endogenous) and responds to the environment (Entrainment (chronobiology), entrained by the environment). These 24-hour rhythms are driven by a circadian clock, and they have been widely observed in animals, plants, fungi and cyanobacteria. The term ''circadian'' comes from the Latin ''wikt:circa#Latin, circa'', meaning "approximately", and ''dies'', meaning "day". Processes with 24-hour cycles are more generally called diurnal rhythms; diurnal rhythms should not be called circadian rhythms unless they can be confirmed as endogenous, and not environmental. Although circadian rhythms are endogenous, they are adjusted to the local environment by external cues called zeitgebers (German for "time givers"), which include light, temperature and redox cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nature (journal)
''Nature'' is a British weekly scientific journal founded and based in London, England. As a multidisciplinary publication, ''Nature'' features peer-reviewed research from a variety of academic disciplines, mainly in science and technology. It has core editorial offices across the United States, continental Europe, and Asia under the international scientific publishing company Springer Nature. ''Nature'' was one of the world's most cited scientific journals by the Science Edition of the 2019 ''Journal Citation Reports'' (with an ascribed impact factor of 42.778), making it one of the world's most-read and most prestigious academic journals. , it claimed an online readership of about three million unique readers per month. Founded in autumn 1869, ''Nature'' was first circulated by Norman Lockyer and Alexander Macmillan as a public forum for scientific innovations. The mid-20th century facilitated an editorial expansion for the journal; ''Nature'' redoubled its efforts in exp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleobase
Nucleobases, also known as ''nitrogenous bases'' or often simply ''bases'', are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA). Five nucleobases—adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called ''primary'' or ''canonical''. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings. In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
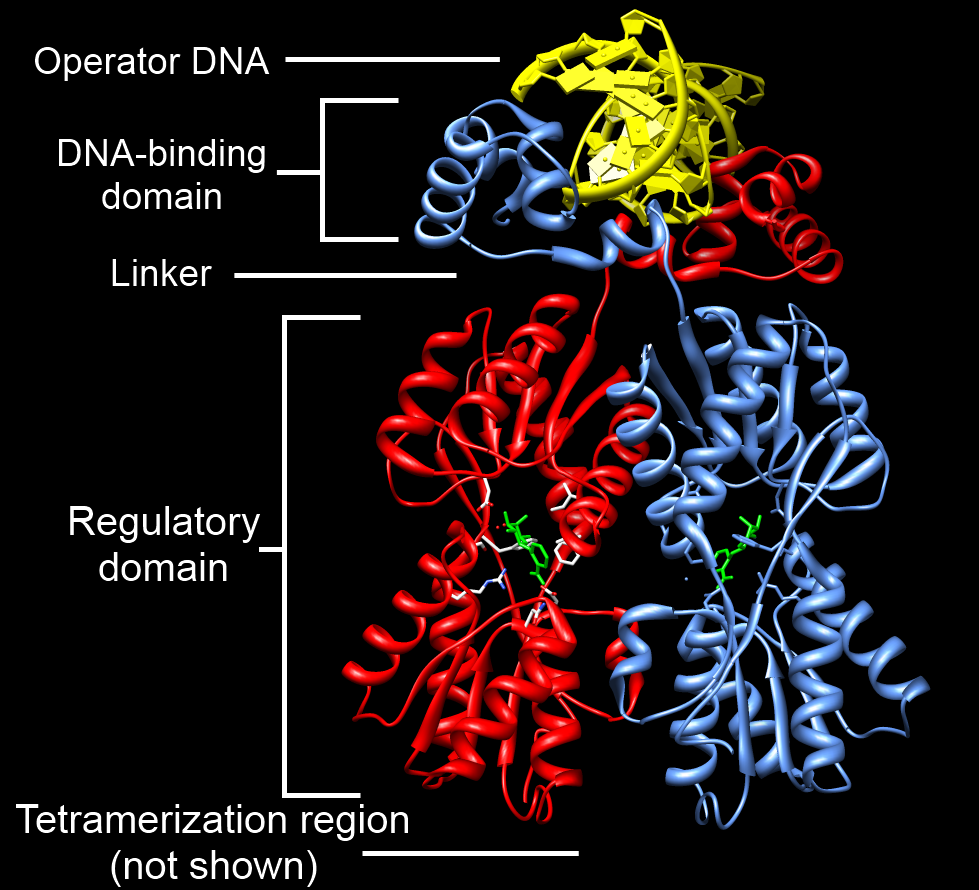
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Channelrhodopsin
Channelrhodopsins are a subfamily of retinylidene proteins ( rhodopsins) that function as light-gated ion channels. They serve as sensory photoreceptors in unicellular green algae, controlling phototaxis: movement in response to light. Expressed in cells of other organisms, they enable light to control electrical excitability, intracellular acidity, calcium influx, and other cellular processes (see optogenetics). Channelrhodopsin-1 (ChR1) and Channelrhodopsin-2 (ChR2) from the model organism ''Chlamydomonas reinhardtii'' are the first discovered channelrhodopsins. Variants that are sensitive to different colors of light or selective for specific ions (ACRs, KCRs) have been cloned from other species of algae and protists. History Phototaxis and photoorientation of microalgae have been studied over more than hundred years in many laboratories worldwide. In 1980, Ken Foster developed the first consistent theory about the functionality of algal eyes. He also analyzed published ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence Microscope
A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image. Principle The specimen is illuminated with light of a specific wavelength (or wavelengths) which is absorbed by the fluorophores, causing them to emit light of longer wavelengths (i.e., of a different color than the absorbed light). The illumination light is separated from the much weaker emitted fluorescence through the use of a spectral emission filter. Typical components of a fluorescence microscope are a light source (xenon arc lamp or mercury-vapor lamp are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Calcium
Calcium is a chemical element with the symbol Ca and atomic number 20. As an alkaline earth metal, calcium is a reactive metal that forms a dark oxide-nitride layer when exposed to air. Its physical and chemical properties are most similar to its heavier homologues strontium and barium. It is the fifth most abundant element in Earth's crust, and the third most abundant metal, after iron and aluminium. The most common calcium compound on Earth is calcium carbonate, found in limestone and the fossilised remnants of early sea life; gypsum, anhydrite, fluorite, and apatite are also sources of calcium. The name derives from Latin ''calx'' "lime", which was obtained from heating limestone. Some calcium compounds were known to the ancients, though their chemistry was unknown until the seventeenth century. Pure calcium was isolated in 1808 via electrolysis of its oxide by Humphry Davy, who named the element. Calcium compounds are widely used in many industries: in foods and pharma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |