|
Floxing
In genetics, floxing refers to the sandwiching of a DNA sequence (which is then said to be floxed) between two '' lox P'' sites. The terms are constructed upon the phrase "flanking/flanked by LoxP". Recombination between LoxP sites is catalysed by Cre recombinase. Floxing a gene allows it to be deleted (knocked out), translocated or inverted in a process called Cre-Lox recombination. The floxing of genes is essential in the development of scientific model systems as it allows researchers to have spatial and temporal alteration of gene expression. Moreover, animals such as mice can be used as models to study human disease. Therefore, Cre-lox system can be used in mice to manipulate gene expression in order to study human diseases and drug development. For example, using the Cre-lox system, researchers can study oncogenes and tumor suppressor genes and their role in development and progression of cancer in mice models. Uses in research Floxing a gene allows it to be deleted (k ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Floxing Flow Chart
In genetics, floxing refers to the sandwiching of a DNA sequence (which is then said to be floxed) between two '' lox P'' sites. The terms are constructed upon the phrase "flanking/flanked by LoxP". Recombination between LoxP sites is catalysed by Cre recombinase. Floxing a gene allows it to be deleted (knocked out), translocated or inverted in a process called Cre-Lox recombination. The floxing of genes is essential in the development of scientific model systems as it allows researchers to have spatial and temporal alteration of gene expression. Moreover, animals such as mice can be used as models to study human disease. Therefore, Cre-lox system can be used in mice to manipulate gene expression in order to study human diseases and drug development. For example, using the Cre-lox system, researchers can study oncogenes and tumor suppressor genes and their role in development and progression of cancer in mice models. Uses in research Floxing a gene allows it to be deleted (k ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code, or its genotype, and the influence of environmental factors. Both factors may interact, further affecting phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book ''The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as cad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures. Discovery Members of the Hsp70 family are very strongly upregulated by heat stress and toxic chemicals, particularly heavy metals such as arsenic, cadmium, copper, mercury, etc. Heat shock was originally discovered by Ferruccio Ritossa in the 1960s when a lab worker accidentally boosted the incubation temperature of Drosophila (fruit flies). When ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand Binding Domain
In the field of molecular biology, nuclear receptors are a class of proteins responsible for sensing steroids, thyroid hormones, vitamins, and certain other molecules. These receptors work with other proteins to regulate the expression of specific genes thereby controlling the development, homeostasis, and metabolism of the organism. Nuclear receptors bind directly to DNA regulating the expression of adjacent genes; hence these receptors are classified as transcription factors. The regulation of gene expression by nuclear receptors often occurs in the presence of a ligand—a molecule that affects the receptor's behavior. Ligand binding to a nuclear receptor results in a conformational change activating the receptor. The result is up- or down-regulation of gene expression. A unique property of nuclear receptors that differentiates them from other classes of receptors is their direct control of genomic DNA. Nuclear receptors play key roles in both embryonic development and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Estrogen Receptor
Estrogen receptors (ERs) are a group of proteins found inside cells. They are receptors that are activated by the hormone estrogen ( 17β-estradiol). Two classes of ER exist: nuclear estrogen receptors (ERα and ERβ), which are members of the nuclear receptor family of intracellular receptors, and membrane estrogen receptors (mERs) (GPER (GPR30), ER-X, and Gq-mER), which are mostly G protein-coupled receptors. This article refers to the former (ER). Once activated by estrogen, the ER is able to translocate into the nucleus and bind to DNA to regulate the activity of different genes (i.e. it is a DNA-binding transcription factor). However, it also has additional functions independent of DNA binding. As hormone receptors for sex steroids (steroid hormone receptors), ERs, androgen receptors (ARs), and progesterone receptors (PRs) are important in sexual maturation and gestation. Proteomics There are two different forms of the estrogen receptor, usually referred to as α a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
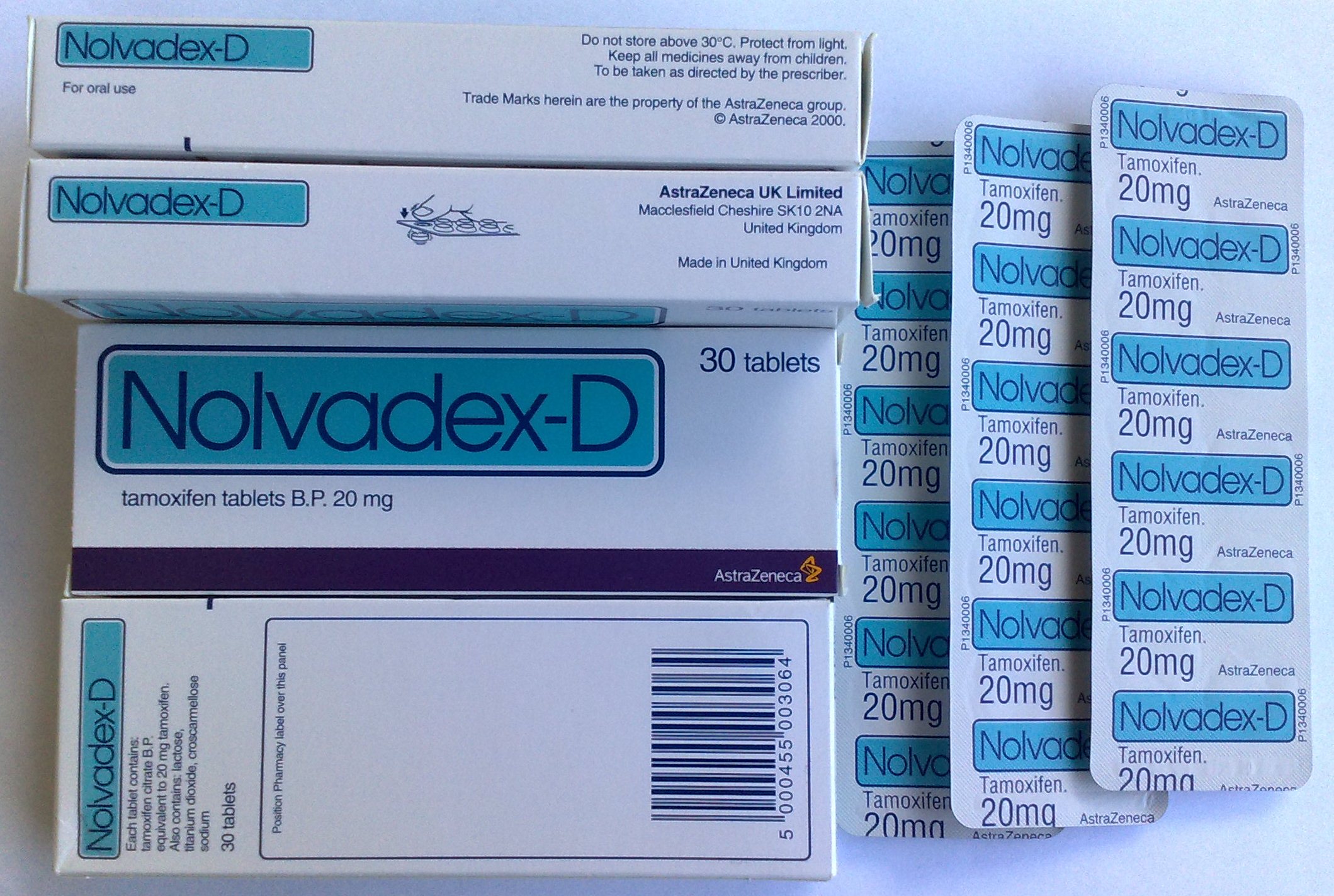
Tamoxifen
Tamoxifen, sold under the brand name Nolvadex among others, is a selective estrogen receptor modulator used to prevent breast cancer in women and treat breast cancer in women and men. It is also being studied for other types of cancer. It has been used for Albright syndrome. Tamoxifen is typically taken daily by mouth for five years for breast cancer. Serious side effects include a small increased risk of uterine cancer, stroke, vision problems, and pulmonary embolism. Common side effects include irregular periods, weight loss, and hot flashes. It may cause harm to the baby if taken during pregnancy or breastfeeding. It is a selective estrogen-receptor modulator (SERM) and works by decreasing the growth of breast cancer cells. It is a member of the triphenylethylene group of compounds. Tamoxifen was initially made in 1962, by chemist Dora Richardson. It is on the World Health Organization's List of Essential Medicines. Tamoxifen is available as a generic medication. In 202 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Myosin Heavy Chain
Myosins () are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility. The first myosin (M2) to be discovered was in 1864 by Wilhelm Kühne. Kühne had extracted a viscous protein from skeletal muscle that he held responsible for keeping the tension state in muscle. He called this protein ''myosin''. The term has been extended to include a group of similar ATPases found in the cells of both striated muscle tissue and smooth muscle tissue. Following the discovery in 1973 of enzymes with myosin-like function in '' Acanthamoeba castellanii'', a global range of divergent myosin genes have been discovered throughout the realm of eukaryotes. Although myosin was originally thought to be restricted to muscle cells (hence '' myo-''(s) + '' -in''), there is no single "myosin"; rather it is a very large superfamily of genes whose prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
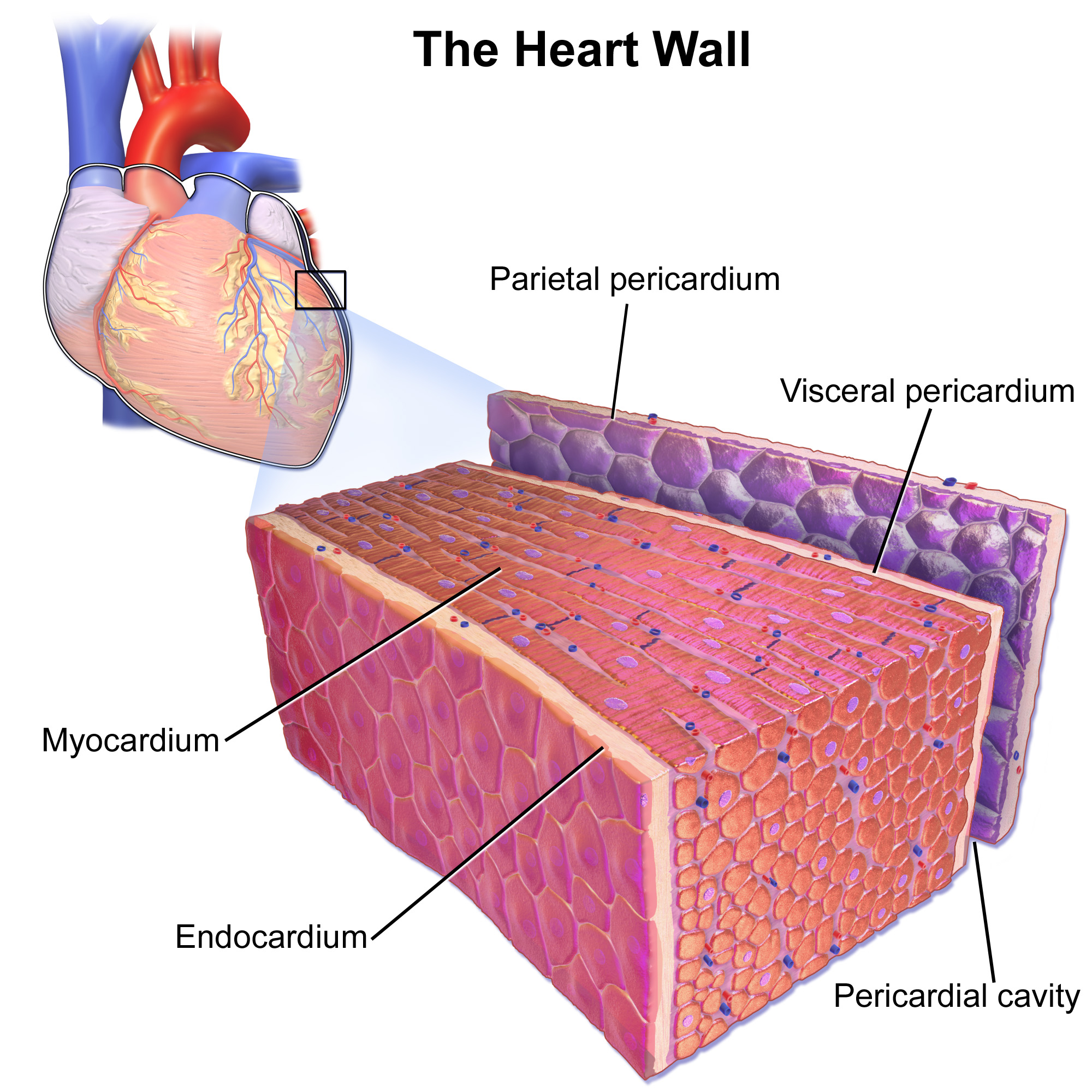
Cardiomyocytes
Cardiac muscle (also called heart muscle, myocardium, cardiomyocytes and cardiac myocytes) is one of three types of vertebrate muscle tissues, with the other two being skeletal muscle and smooth muscle. It is an involuntary, striated muscle that constitutes the main tissue of the wall of the heart. The cardiac muscle (myocardium) forms a thick middle layer between the outer layer of the heart wall (the pericardium) and the inner layer (the endocardium), with blood supplied via the coronary circulation. It is composed of individual cardiac muscle cells joined by intercalated discs, and encased by collagen fibers and other substances that form the extracellular matrix. Cardiac muscle contracts in a similar manner to skeletal muscle, although with some important differences. Electrical stimulation in the form of a cardiac action potential triggers the release of calcium from the cell's internal calcium store, the sarcoplasmic reticulum. The rise in calcium causes the cell's my ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FLP-FRT Recombination
In genetics, Flp-''FRT'' recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions ''in vivo''. It is analogous to Cre-''lox'' recombination but involves the recombination of sequences between short flippase recognition target (''FRT'') sites by the recombinase flippase (''Flp'') derived from the 2 µ plasmid of baker's yeast ''Saccharomyces cerevisiae''. The 34bp minimal FRT site sequence has the sequence ::5'3' for which flippase (Flp) binds to both 13-bp 5'-GAAGTTCCTATTC-3' arms flanking the 8 bp spacer, i.e. the site-specific recombination (region of crossover) in reverse orientation. ''FRT''-mediated cleavage occurs just ahead from the asymmetric 8bp core region (5''3') on the top strand and behind this sequence on the bottom strand. Several variant ''FRT'' sites exist, but recombination can usually occur only between two ''identical'' ''FRT''s but generally not among ''non-identical'' (" ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cre-Lox Recombination
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community. The system consists of a single enzyme, Cre recombinase, that recombines a pair of short target sequences called the ''Lox'' sequences. This system can be implemented without inserting any extra supporting proteins or sequences. The Cre enzyme and the original ''Lox'' sit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conditional Gene Knockout
Conditional gene knockout is a technique used to eliminate a specific gene in a certain tissue, such as the liver. This technique is useful to study the role of individual genes in living organisms. It differs from traditional gene knockout because it targets specific genes at specific times rather than being deleted from beginning of life. Using the conditional gene knockout technique eliminates many of the side effects from traditional gene knockout. In traditional gene knockout, embryonic death from a gene mutation can occur, and this prevents scientists from studying the gene in adults. Some tissues cannot be studied properly in isolation, so the gene must be inactive in a certain tissue while remaining active in others. With this technology, scientists are able to knockout genes at a specific stage in development and study how the knockout of a gene in one tissue affects the same gene in other tissues. Technique The most commonly used technique is the Cre-lox recombination syst ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |