|
Epigenome Editing
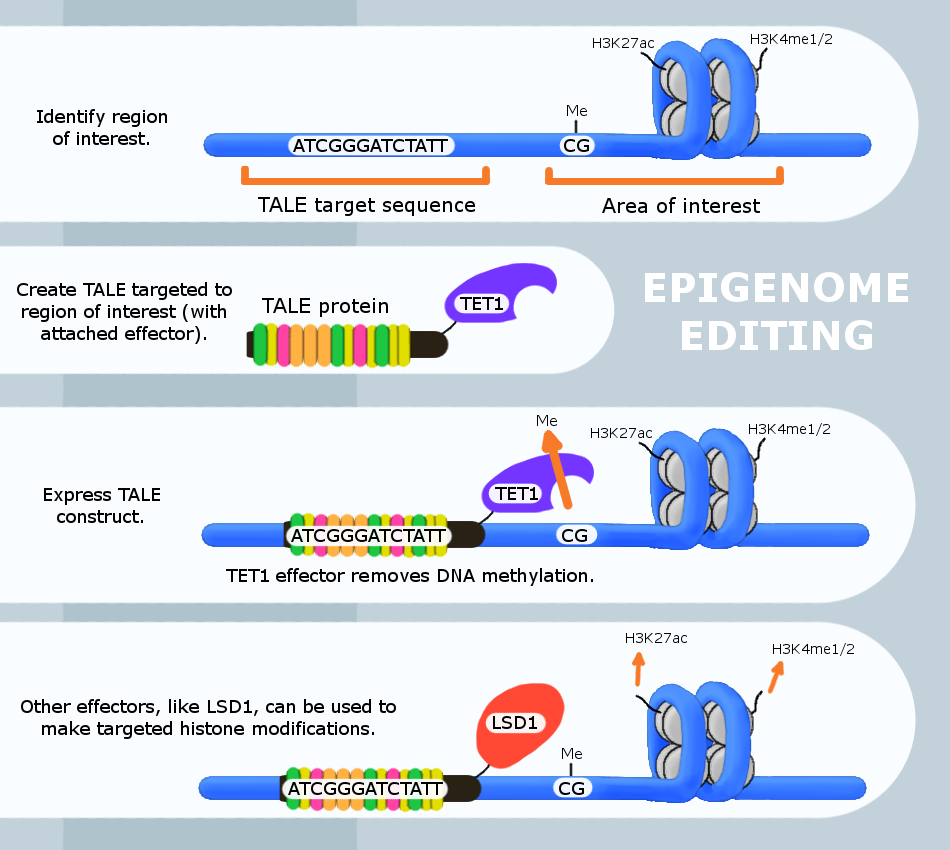
Epigenome editing or Epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites (as opposed to whole-genome modifications). Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question. The engineered proteins used for epigenome editing are composed of a DNA binding domain that target specific sequences and an effector domain that modifies epigenomic features. Currently, three major groups of DNA binding proteins have been predominantly used for epigenome editing: Zinc finger proteins, Transcription Activator-Like Effectors (TALEs) and nuclease deficient Cas9 fusions (CRISPR). Gener ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenome Editing
Epigenome editing or Epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites (as opposed to whole-genome modifications). Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question. The engineered proteins used for epigenome editing are composed of a DNA binding domain that target specific sequences and an effector domain that modifies epigenomic features. Currently, three major groups of DNA binding proteins have been predominantly used for epigenome editing: Zinc finger proteins, Transcription Activator-Like Effectors (TALEs) and nuclease deficient Cas9 fusions (CRISPR). Gener ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRY2
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name ''cryptochrome'' was proposed as a ''portmanteau'' combining the ''chromatic'' nature of the photoreceptor, and the ''cryptogamic'' organisms on which many blue-light studies were carried out. The two genes ''Cry1'' and ''Cry2'' code the two cryptochrome proteins CRY1 and CRY2. In insects and plants, CRY1 regulates the circadian clock in a light-dependent fashion, whereas in mammals, CRY1 and CRY2 act as light-independent inhibitors of CLOCK-BMAL1 components of the circadian clock. In plants, blue-light photoreception can be used to cue developmental signals. Besides chlorophylls, cryptochromes are the only proteins known to form photoinduced radical-pairs ''in vivo''. These appear to enable some animals ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cryptochrome
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name ''cryptochrome'' was proposed as a ''portmanteau'' combining the '' chromatic'' nature of the photoreceptor, and the ''cryptogamic'' organisms on which many blue-light studies were carried out. The two genes ''Cry1'' and ''Cry2'' code the two cryptochrome proteins CRY1 and CRY2. In insects and plants, CRY1 regulates the circadian clock in a light-dependent fashion, whereas in mammals, CRY1 and CRY2 act as light-independent inhibitors of CLOCK-BMAL1 components of the circadian clock. In plants, blue-light photoreception can be used to cue developmental signals. Besides chlorophylls, cryptochromes are the only proteins known to form photoinduced radical-pairs ''in vivo''. These appear to enable some animal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histones
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histones ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K4me1
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono-methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers. Nomenclature H3K4me1 indicates monomethylation of lysine 4 on histone H3 protein subunit: Lysine methylation This diagram shows the progressive methylation of a lysine residue. The mono-methylation denotes the methylation present in H3K4me1. Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The carboxyl (C) terminal end of these histones contribute to his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CpG Sites
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands (or CG islands). Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Methylated cytosines often mutate to thymines. In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a CpG island. CpG characteristics Definition ''CpG'' is shorthand for ''5'—C—phosphate—G—3' '', that is, cytosine and guanine separated by only one phosphate group; phosphate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EP300
Histone acetyltransferase p300 also known as p300 HAT or E1A-associated protein p300 (where E1A = adenovirus early region 1A) also known as EP300 or p300 is an enzyme that, in humans, is encoded by the ''EP300'' gene. It functions as histone acetyltransferase that regulates transcription of genes via chromatin remodeling by allowing histone proteins to wrap DNA less tightly. This enzyme plays an essential role in regulating cell growth and division, prompting cells to mature and assume specialized functions (differentiate), and preventing the growth of cancerous tumors. The p300 protein appears to be critical for normal development before and after birth. The EP300 gene is located on the long (q) arm of the human chromosome 22 at position 13.2. This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. EP300 is closely related to another gene, CREB binding protein, which is found on human chromosome 16. Function p300 HAT functions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA (cytosine-5)-methyltransferase 3A
DNA (cytosine-5)-methyltransferase 3A is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene. This enzyme is responsible for '' de novo'' DNA methylation. Such function is to be distinguished from maintenance DNA methylation which ensures the fidelity of replication of inherited epigenetic patterns. DNMT3A forms part of the family of DNA methyltransferase enzymes, which consists of the protagonists DNMT1, DNMT3A and DNMT3B. While ''de novo'' DNA methylation modifies the information passed on by the parent to the progeny, it enables key epigenetic modifications essential for processes such as cellular differentiation and embryonic development, transcriptional regulation, heterochromatin formation, X-inactivation, imprinting and genome stability. DNMT3a is the gene most commonly found mutated in clonal hematopoiesis, a common aging-related phenomenon in which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
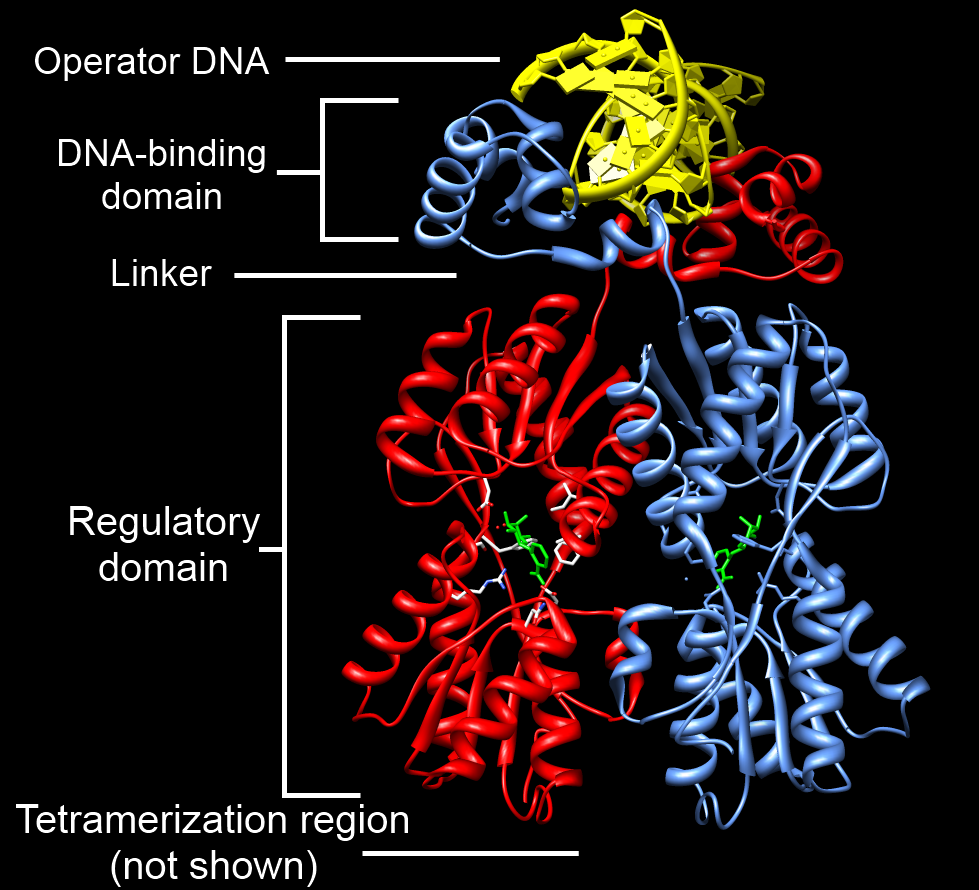
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. ''Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, there are a number of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CIB1
Calcium and integrin-binding protein 1 is a protein that in humans is encoded by the ''CIB1'' gene. The protein encoded by this gene is a member of the calcium-binding protein family. The specific function of this protein has not yet been determined; however this protein is known to interact with DNA-dependent protein kinase and may play a role in kinase-phosphatase regulation of DNA end-joining. This protein also interacts with integrin alpha(IIb)beta(3), which may implicate this protein as a regulatory molecule for alpha(IIb)beta(3). Interactions CIB1 has been shown to interact with RAC3, PSEN2, DNA-PKcs, UBR5 and CD61 Integrin beta-3 (β3) or CD61 is a protein that in humans is encoded by the ''ITGB3'' gene. CD61 is a cluster of differentiation found on thrombocytes. Structure and function The ITGB3 protein product is the integrin beta chain beta 3. Integrins .... References Further reading * * * * * * * * * * * * * * * * * * External links * EF-hand-containin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LSD1
Lysine-specific histone demethylase 1A (LSD1) also known as lysine (K)-specific demethylase 1A (KDM1A) is a protein in humans that is encoded by the KDM1A gene. LSD1 is a flavin-dependent monoamine oxidase, which can demethylate mono- and di-methylated lysines, specifically histone 3, lysines 4 and 9 (H3K4 and H3K9). This enzyme can have roles critical in embryogenesis and tissue-specific differentiation, as well as oocyte growth. KDM1A was the first histone demethylase to be discovered though more than 30 have been described. Structure This gene encodes a nuclear protein containing a SWIRM domain, a FAD-binding motif, and an amine oxidase domain. This protein is a component of several histone deacetylase complexes, though it silences genes by functioning as a histone demethylase. Function LSD1 (lysine-specific demethylase 1), also known as KDM1, is the first of several protein lysine demethylases discovered. Through a FAD-dependent oxidative reaction, LSD1 specifical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |