Epigenome Editing on:
[Wikipedia]
[Google]
[Amazon]
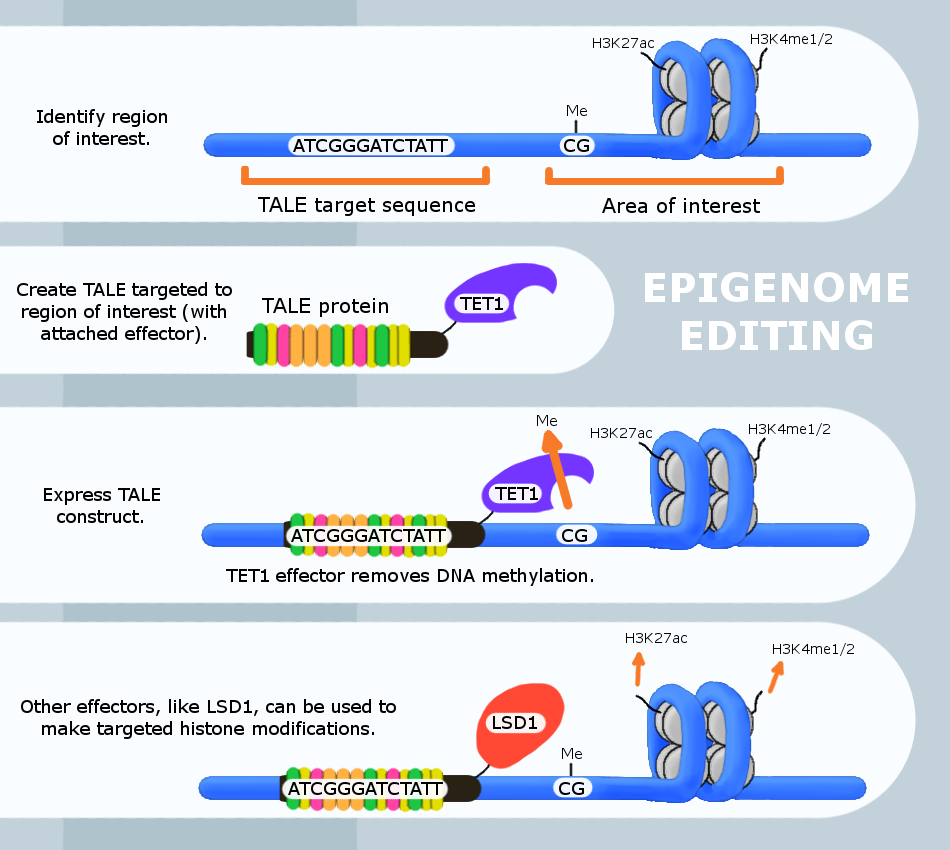 Epigenome editing or Epigenome engineering is a type of genetic engineering in which the
Epigenome editing or Epigenome engineering is a type of genetic engineering in which the
CRISPR Activation of Single Genes Turns Skin Cells to Stem Cells
* * {{refend Epigenetics Genetic engineering Genome editing
 Epigenome editing or Epigenome engineering is a type of genetic engineering in which the
Epigenome editing or Epigenome engineering is a type of genetic engineering in which the epigenome
An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism; these changes can be passed down to an organism's offspring via transgenerational stranded epigenetic inheritance. Changes to the epigenome ...
is modified at specific sites using engineered molecules targeted to those sites (as opposed to whole-genome modifications). Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question.
The engineered proteins used for epigenome editing are composed of a DNA binding domain that target specific sequences and an effector domain that modifies epigenomic features. Currently, three major groups of DNA binding proteins have been predominantly used for epigenome editing: Zinc finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized struct ...
proteins, Transcription Activator-Like Effectors (TALEs) and nuclease deficient Cas9 fusions (CRISPR
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bac ...
).
General concept
Comparing genome-wideepigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are " ...
maps with gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. ...
has allowed researchers to assign either activating or repressing roles to specific modifications. The importance of DNA sequence in regulating the epigenome has been demonstrated by using DNA motifs to predict epigenomic modification. Further insights into mechanisms behind epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are " ...
have come from in vitro
''In vitro'' (meaning in glass, or ''in the glass'') studies are performed with microorganisms, cells, or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in biology and ...
biochemical and structural analyses. Using model organisms, researchers have been able to describe the role of many chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
factors through knockout
A knockout (abbreviated to KO or K.O.) is a fight-ending, winning criterion in several full-contact combat sports, such as boxing, kickboxing, muay thai, mixed martial arts, karate, some forms of taekwondo and other sports involving striking, a ...
studies. However knocking out an entire chromatin modifier has massive effects on the entire genome, which may not be an accurate representation of its function in a specific context. As one example of this, DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts ...
occurs at repeat regions, promoters, enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, and gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
bodies. Although DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts ...
at gene promoters typically correlates with gene repression, methylation at gene bodies is correlated with gene activation, and DNA methylation may also play a role in gene splicing. The ability to directly target and edit individual methylation sites is critical to determining the exact function of DNA methylation at a specific site. Epigenome editing is a powerful tool that allows this type of analysis. For site-specific DNA methylation editing as well as for histone editing, genome editing systems have been adapted into epigene editing systems. In short, genome homing proteins with engineered or naturally occurring nuclease functions for gene editing, can be mutated and adapted into purely delivery systems. An epigenetic modifying enzyme or domain can be fused to the homing protein and local epigenetic modifications can be altered upon protein recruitment.
Targeting proteins
TALE
The Transcription Activator-Like Effector (TALE) protein recognizes specific DNA sequences based on the composition of its DNA binding domain. This allows the researcher to construct different TALE proteins to recognize a target DNA sequence by editing the TALE's primary protein structure. The binding specificity of this protein is then typically confirmed usingChromatin Immunoprecipitation Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genom ...
(ChIP) and Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frede ...
of the resulting DNA fragment. This confirmation is still required on all TALE sequence recognition research.
When used for epigenome editing, these DNA binding proteins are attached to an effector protein. Effector proteins that have been used for this purpose include Ten-eleven translocation methylcytosine dioxygenase 1 (TET1), Lysine (K)-specific demethylase 1A (LSD1) and Calcium and integrin binding protein 1 (CIB1).
Zinc finger proteins
The use ofzinc finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized struct ...
-fusion proteins to recognize sites for epigenome editing has been explored as well. Maeder et al. has constructed a ZF-TET1 protein for use in DNA demethylation. These zinc finger proteins work similarly to TALE proteins in that they are able to bind to sequence specific sites in on the DNA based on their protein structure which can be modified. Chen et al. have successfully used a zinc finger DNA binding domain coupled with the TET1
Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes, in humans it is encoded by the TET1 gene. Its function, regulation, and utilizable pathways remain a matter of current research while it seems t ...
protein to induce demethylation of several previously silenced genes. Kungulovski and Jeltsch successfully used ZFP-guided deposition of DNA methylation gene to cause gene silencing but the DNA methylation and silencing were lost when the trigger signal stopped. The authors suggest for stable epigenetic changes, there must be either multiple depositions of DNA methylation of related epigenetic marks, or long-lasting trigger stimuli. ZFP epigenetic editing has shown potential to treat various neurodegenerative diseases.
CRISPR-Cas
The Clustered Regulatory Interspaced Short Palindromic Repeat (CRISPR)-Cas system functions as a DNA site-specific nuclease. In the well-studied type II CRISPR system, the Cas9 nuclease associates with a chimera composed of tracrRNA and crRNA. This chimera is frequently referred to as a guide RNA (gRNA). When the Cas9 protein associates with a DNA region-specific gRNA, the Cas9 cleaves DNA at targeted DNA loci. However, when the D10A and H840A point mutations are introduced, a catalytically-dead Cas9 (dCas9) is generated that can bind DNA but will not cleave. The dCas9 system has been utilized for targeted epigenetic reprogramming in order to introduce site-specific DNA methylation. By fusing theDNMT3a
DNA (cytosine-5)-methyltransferase 3A is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.
This enzyme is responsible f ...
catalytic domain with the dCas9 protein, dCas9-DNMT3a is capable of achieving targeted DNA methylation of a targeted region as specified by the present guide RNA. Similarly, dCas9 has been fused with the catalytic core of the human acetyltransferase p300. dCas9-p300 successfully catalyzes targeted acetylation of histone H3 lysine 27.
A variant in CRISPR epigenome editing (called FIRE-Cas9) allows to reverse the changes made, in case something went wrong.
''CRISPRoff'' is a dead Cas9 fusion protein that can be used to heritably silence the gene expression of "most genes" and allows for reversible modifications.
Commonly used effector proteins
TET1
Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes, in humans it is encoded by the TET1 gene. Its function, regulation, and utilizable pathways remain a matter of current research while it seems t ...
induces demethylation of cytosine at CpG sites
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
. This protein has been used to activate genes that are repressed by CpG methylation and to determine the role of individual CpG methylation sites. LSD1 induces the demethylation of H3K4me1
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono- methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers.
Nomenclature
H3K4m ...
/2, which also causes an indirect effect of deacetylation on H3K27. This effector can be used on histones
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are ...
in enhancer regions, which can changes the expression of neighboring genes. CIB1
Calcium and integrin-binding protein 1 is a protein that in humans is encoded by the ''CIB1'' gene.
The protein encoded by this gene is a member of the calcium-binding protein family. The specific function of this protein has not yet been determi ...
is a light sensitive cryptochrome
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields ...
, this cryptochrome is fused to the TALE protein. A second protein contains an interaction partner (CRY2
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields ...
) fused with a chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
/DNA modifier (ex. SID4X). CRY2
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields ...
is able to interact with CIB1
Calcium and integrin-binding protein 1 is a protein that in humans is encoded by the ''CIB1'' gene.
The protein encoded by this gene is a member of the calcium-binding protein family. The specific function of this protein has not yet been determi ...
when the cryptochrome has been activated by illumination with blue light. The interaction allows the chromatin modifier to act on the desired location. This means that the modification can be performed in an inducible and reversible manner, which reduces long-term secondary effects that would be caused by constitutive epigenetic modification.
Applications
Studying enhancer function and activity
Editing of gene enhancer regions in the genome through targeted epigenetic modification has been demonstrated by Mendenhall et al. (2013). This study utilized a TALE-LSD1 effector fusion protein in order to target enhancers of genes, to induce enhancer silencing in order to deduce enhancer activity and gene control. Targeting specific enhancers followed by locus specific RT-qPCR allows for the genes affected by the silenced enhancer to be determined. Alternatively, inducing enhancer silencing in regions upstream of genes allows for gene expression to be altered. RT-qPCR can then be utilized to study effects of this on gene expression. This allows for enhancer function and activity to be studied in detail.Determining the function of specific methylation sites
It is important to understand the role specific methylation sites play regulating in gene expression. To study this, one research group used a TALE-TET1 fusion protein to demethylate a single CpG methylation site. Although this approach requires many controls to ensure specific binding to target loci, a properly performed study using this approach can determine the biological function of a specific CpG methylation site.Determining the role of epigenetic modifications directly
Epigenetic editing using an inducible mechanism offers a wide array of potential use to study epigenetic effects in various states. One research group employed an optogenetic two-hybrid system which integrated the sequence specific TALE DNA-binding domain with a light-sensitive cryptochrome 2 protein (CIB1
Calcium and integrin-binding protein 1 is a protein that in humans is encoded by the ''CIB1'' gene.
The protein encoded by this gene is a member of the calcium-binding protein family. The specific function of this protein has not yet been determi ...
). Once expressed in the cells, the system was able to inducibly edit histone modifications and determine their function in a specific context.
Functional engineering
Targeted regulation of disease-related genes may enable novel therapies for many diseases, especially in cases where adequate gene therapies are not yet developed or are inappropriate. While transgenerational and population level consequences are not fully understood, it may become a major tool for applied functional genomics andpersonalized medicine
Personalized medicine, also referred to as precision medicine, is a medical model that separates people into different groups—with medical decisions, practices, interventions and/or products being tailored to the individual patient based on the ...
. As with RNA editing
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms ...
, it does not involve genetic changes and their accompanying risks. One example of a potential functional use of epigenome editing was described in 2021: repressing Nav1.7 gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. ...
via CRISPR-dCas9 which showed therapeutic potential in three mouse models of chronic pain.
In 2022, research assessed its usefulness in reducing tau protein
The tau proteins (abbreviated from tubulin associated unit) are a group of six highly soluble protein isoforms produced by alternative splicing from the gene ''MAPT'' (microtubule-associated protein tau). They have roles primarily in maintaining ...
levels, regulating a protein involved in Huntington's disease
Huntington's disease (HD), also known as Huntington's chorea, is a neurodegenerative disease that is mostly inherited. The earliest symptoms are often subtle problems with mood or mental abilities. A general lack of coordination and an uns ...
, targeting an inherited form of obesity, and Dravet syndrome.
Limitations
Sequence specificity is critically important in epigenome editing and must be carefully verified (this can be done usingchromatin immunoprecipitation Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genom ...
followed by Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frede ...
to verify the targeted sequence). It is unknown if the TALE fusion may cause effects on the catalytic activity of the epigenome modifier. This could be especially important in effector proteins that require multiple subunits and complexes such as the Polycomb
Polycomb-group proteins (PcG proteins) are a family of protein complexes first discovered in fruit flies that can remodel chromatin such that epigenetic silencing of genes takes place. Polycomb-group proteins are well known for silencing Hox gene ...
repressive complex. Proteins used for epigenome editing may obstruct ligands and substrates at the target site. The TALE protein itself may even compete with transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The f ...
if they are targeted to the same sequence. In addition, DNA repair systems could reverse the alterations on the chromatin and prevent the desired changes from being made. It is therefore necessary for fusion constructs and targeting mechanisms to be optimized for reliable and repeatable epigenome editing.
See also
* DNA editing *RNA editing
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms ...
References
Further reading
* * * * * * * * * * * * *Tompkins JD. www.epigenomeengineering.comCRISPR Activation of Single Genes Turns Skin Cells to Stem Cells
* * {{refend Epigenetics Genetic engineering Genome editing