|
Exchanger (protein)
An antiporter (also called exchanger or counter-transporter) is an integral membrane protein that uses secondary active transport to move two or more molecules in opposite directions across a phospholipid membrane. It is a type of cotransporter, which means that uses the energetically favorable movement of one molecule down its electrochemical gradient to power the energetically unfavorable movement of another molecule up its electrochemical gradient. This is in contrast to symporters, which are another type of cotransporter that moves two or more ions in the same direction, and primary active transport, which is directly powered by ATP. Transport may involve one or more of each type of solute. For example, the Na+/Ca2+ exchanger, found in the plasma membrane of many cells, moves three sodium ions in one direction, and one calcium ion in the other. As with sodium in this example, antiporters rely on an established gradient that makes entry of one ion energetically favorable t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Transport Proteins
Active may refer to: Music * Active (album), ''Active'' (album), a 1992 album by Casiopea * Active (song), "Active" (song), a 2024 song by Asake and Travis Scott from Asake's album ''Lungu Boy'' * Active Records, a record label Ships * Active (ship), ''Active'' (ship), several commercial ships by that name * HMS Active, HMS ''Active'', the name of various ships of the British Royal Navy * USCS Active, USCS ''Active'', a US Coast Survey ship in commission from 1852 to 1861 * USCGC Active, USCGC ''Active'', the name of various ships of the US Coast Guard * USRC Active, USRC ''Active'', the name of various ships of the US Revenue Cutter Service * USS Active, USS ''Active'', the name of various ships of the US Navy Computers and electronics * Active Enterprises, a defunct video game developer * Sky Active, the brand name for interactive features on Sky Digital available in the UK and Ireland * Active (software), software used for open publishing by Indymedia; see Independent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uniporters
Facilitated diffusion (also known as facilitated transport or passive-mediated transport) is the process of spontaneous passive transport (as opposed to active transport) of molecules or ions across a biological membrane via specific transmembrane integral proteins. Being passive, facilitated transport does not directly require chemical energy from ATP hydrolysis in the transport step itself; rather, molecules and ions move down their concentration gradient according to the principles of diffusion. Facilitated diffusion differs from simple diffusion in several ways: # The transport relies on molecular binding between the cargo and the membrane-embedded channel or carrier protein. # The rate of facilitated diffusion is saturable with respect to the concentration difference between the two phases; unlike free diffusion which is linear in the concentration difference. # The temperature dependence of facilitated transport is substantially different due to the presence of an acti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophile
A hydrophile is a molecule or other molecular entity that is intermolecular force, attracted to water molecules and tends to be dissolution (chemistry), dissolved by water.Liddell, H.G. & Scott, R. (1940). ''A Greek-English Lexicon'' Oxford: Clarendon Press. In contrast, hydrophobes are not attracted to water and may seem to be repelled by it. Hygroscopics ''are'' attracted to water, but are not dissolved by water. Molecules A hydrophilic molecule or portion of a molecule is one whose interactions with water and other polar substances are more Thermodynamics, thermodynamically favorable than their interactions with oil or other hydrophobic solvents. They are typically charge-polarized and capable of hydrogen bonding. This makes these molecules soluble not only in water but also in Solvent#Solvent classifications, polar solvents. Hydrophilic molecules (and portions of molecules) can be contrasted with hydrophobic molecules (and portions of molecules). In some cases, both hydrophi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lipid Bilayer
The lipid bilayer (or phospholipid bilayer) is a thin polar membrane made of two layers of lipid molecules. These membranes form a continuous barrier around all cell (biology), cells. The cell membranes of almost all organisms and many viruses are made of a lipid bilayer, as are the nuclear envelope, nuclear membrane surrounding the cell nucleus, and biological membrane, membranes of the membrane-bound organelles in the cell. The lipid bilayer is the barrier that keeps ions, proteins and other molecules where they are needed and prevents them from diffusing into areas where they should not be. Lipid bilayers are ideally suited to this role, even though they are only a few nanometers in width, because they are impermeable to most water-soluble (hydrophilic) molecules. Bilayers are particularly impermeable to ions, which allows cells to regulate salt concentrations and pH by transporting ions across their membranes using proteins called Ion transporter, ion pumps. Biological bilaye ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Region
Cell surface receptors (membrane receptors, transmembrane receptors) are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving (binding to) extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the extracellular space. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane. Structure and mechanism Many membrane receptors are transmembrane proteins. There are various kinds, including glycoproteins and lipoproteins. Hundreds of different receptors are known and many more have yet to be studied. Transmembrane receptors are typically classified based on their tertiary (three-dimensional) struc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand (biochemistry)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from Latin ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
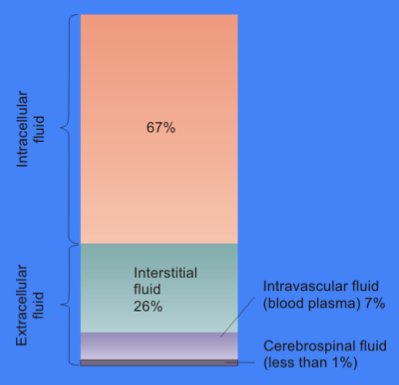
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and proper ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Extracellular Space
Extracellular space refers to the part of a multicellular organism outside the cells, usually taken to be outside the plasma membranes, and occupied by fluid. This is distinguished from intracellular space, which is inside the cells. The composition of the extracellular space includes metabolites, ions, proteins, and many other substances that might affect cellular function. For example, neurotransmitters "jump" from cell to cell to facilitate the transmission of an electric current in the nervous system. Hormones also act by travelling the extracellular space towards cell receptors. In cell biology, molecular biology and related fields, the word extracellular (or sometimes extracellular space) means "outside the cell". This space is usually taken to be outside the plasma membranes, and occupied by fluid (see extracellular matrix). The term is used in contrast to intracellular (inside the cell). According to the Gene Ontology, the extracellular space is a cellular component def ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors. A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a ''conformational change''. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. Laboratory analysis Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macromo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATP Hydrolysis
ATP hydrolysis is the catabolic reaction process by which chemical energy that has been stored in the high-energy phosphoanhydride bonds in adenosine triphosphate (ATP) is released after splitting these bonds, for example in muscles, by producing work in the form of mechanical energy. The product is adenosine diphosphate (ADP) and an inorganic phosphate (Pi). ADP can be further hydrolyzed to give energy, adenosine monophosphate (AMP), and another inorganic phosphate (Pi). ATP hydrolysis is the final link between the energy derived from food or sunlight and useful work such as muscle contraction, the establishment of electrochemical gradients across membranes, and biosynthetic processes necessary to maintain life. Anhydridic bonds are often labelled as "''high-energy bonds"''. P-O bonds are in fact fairly strong (~30 kJ/mol stronger than C-N bonds) Darwent, B. deB. (1970). "Bond Dissociation Energies in Simple Molecules", Nat. Stand. Ref. Data Ser., Nat. Bur. Stand. (U.S.) 31, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium–potassium Pump
The sodium–potassium pump (sodium–potassium adenosine triphosphatase, also known as -ATPase, pump, or sodium–potassium ATPase) is an enzyme (an electrogenic transmembrane ATPase) found in the membrane of all animal cells. It performs several functions in cell physiology. The -ATPase enzyme is active (i.e. it uses energy from ATP). For every ATP molecule that the pump uses, three sodium ions are exported and two potassium ions are imported. Thus, there is a net export of a single positive charge per pump cycle. The net effect is an extracellular concentration of sodium ions which is 5 times the intracellular concentration, and an intracellular concentration of potassium ions which is 30 times the extracellular concentration. The sodium–potassium pump was discovered in 1957 by the Danish scientist Jens Christian Skou, who was awarded a Nobel Prize for his work in 1997. Its discovery marked an important step forward in the understanding of how ions get into ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |