|
Endoribonuclease XendoU
In molecular biology, Endoribonuclease XendoU refers to a protein domain. This particular entry represents endoribonucleases involved in RNA biosynthesis which has been named XendoU in ''Xenopus laevis'' (African clawed frog). This protein domain belongs to a family of evolutionarily related proteins. XendoU is a U-specific metal dependent enzyme that produces products with a 2' -3' cyclic phosphate termini. Function The endonuclease, XendoU, is highly involved in the biosynthesis of a specific subclass of ''Xenopus laevis'' encoded small nucleolar RNAs (snoRNA) which are a large family of non-coding RNAs with essential roles in ribosome biogenesis. Most snoRNAs are encoded in introns and are released through the splicing reaction. Others, instead, produced by an alternative pathway consisting of endonucleolytic processing of pre-mRNA. XendoU, is the endoribonuclease responsible for this activity. The XendoU-RNA complex is manganese (Mn2+)-independent. This infers that RNA b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Splicing Reaction
Splice may refer to: Connections * Rope splicing, joining two pieces of rope or cable by weaving the strands of each into the other ** Eye splice, a method of creating a permanent loop in the end of multi stranded rope by means of rope splicing * Splice joint, a method of joining two members end to end in woodworking * Tape splice, the joining of audio tape * Film splice, the joining of film stock * Electrical splice, the joining of wires in electrical wiring * Optical splice, the joining of optical fibers: ** Fusion splicing, a permanent splice between two fibers ** Mechanical splice, a temporary splice between two fibers * Pile splice, connecting two concrete, timber or steel piles for a deep foundation * Line splice, a splice of two cables in the telecommunication industry Genetics * RNA splicing, a natural modification of ribonucleic acids * Genetic engineering, also known as ''gene splicing'', artificially joining pieces of genetic material * Protein splicing, a natural proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteases
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine proteases - ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology(biology)
Homology may refer to: Sciences Biology *Homology (biology), any characteristic of biological organisms that is derived from a common ancestor *Sequence homology, biological homology between DNA, RNA, or protein sequences *Homologous chromosomes, chromosomes in a biological cell that pair up (synapse) during meiosis *Homologous recombination, genetic recombination in which nucleotide sequences are exchanged between molecules of DNA *Homologous desensitization, a receptor decreases its response to a signalling molecule when that agonist is in high concentration *Homology modeling, a method of protein structure prediction Chemistry *Homology (chemistry), the relationship between compounds in a homologous series *Homologous series, a series of organic compounds having different quantities of a repeated unit *Homologous temperature, the temperature of a material as a fraction of its absolute melting point *Homologation reaction, a chemical reaction which produces the next lo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheets
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined version was p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiparallel (biochemistry)
In biochemistry, two biopolymers are antiparallel if they run parallel to each other but with opposite directionality (alignments). An example is the two complementary strands of a DNA double helix, which run in opposite directions alongside each other. Nucleic acids Nucleic acid molecules have a phosphoryl (5') end and a hydroxyl (3') end. This notation follows from organic chemistry nomenclature, and can be used to define the movement of enzymes such as DNA polymerases relative to the DNA strand in a non-arbitrary manner. G-quadruplexes G-quadruplexes, also known as G4 DNA are secondary structures found in nucleic acids that are rich in guanine. These structures are normally located at the telomeres (the ends of the chromosomes). The G-quadruplex can either be parallel or antiparallel depending on the loop configuration, which is a component of the structure. If all the DNA strands run in the same direction, it is termed to be a parallel quadruplex, and is known as a str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Manganese
Manganese is a chemical element with the symbol Mn and atomic number 25. It is a hard, brittle, silvery metal, often found in minerals in combination with iron. Manganese is a transition metal with a multifaceted array of industrial alloy uses, particularly in stainless steels. It improves strength, workability, and resistance to wear. Manganese oxide is used as an oxidising agent; as a rubber additive; and in glass making, fertilisers, and ceramics. Manganese sulfate can be used as a fungicide. Manganese is also an essential human dietary element, important in macronutrient metabolism, bone formation, and free radical defense systems. It is a critical component in dozens of proteins and enzymes. It is found mostly in the bones, but also the liver, kidneys, and brain. In the human brain, the manganese is bound to manganese metalloproteins, most notably glutamine synthetase in astrocytes. Manganese was first isolated in 1774. It is familiar in the laboratory in the form of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


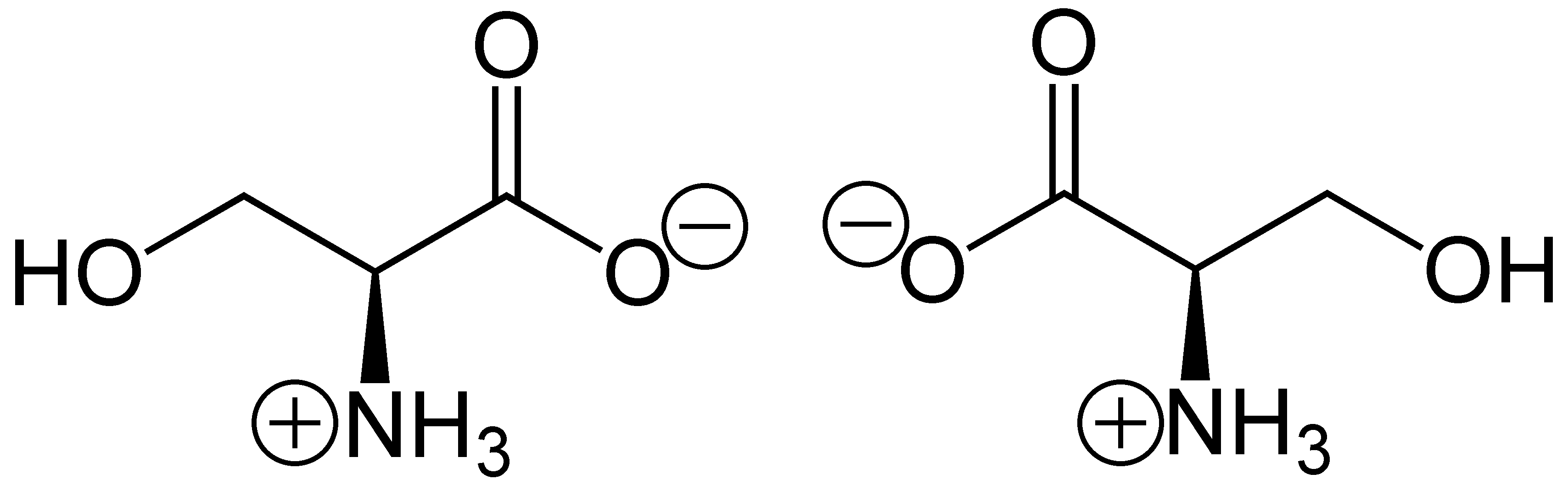

_structure.png)
