|
Death-inducing Signaling Complex
The death-inducing signaling complex or DISC is a multi-protein complex formed by members of the death receptor family of apoptosis-inducing cellular receptors. A typical example is FasR, which forms the DISC upon trimerization as a result of its ligand (FasL) binding. The DISC is composed of the death receptor, FADD, and caspase 8. It transduces a downstream signal cascade resulting in apoptosis. Description The Fas ligands, or cytotoxicity-dependent APO-1-associated proteins, physically associate with APO-1 (also known as the Fas receptor, or CD95), a tumor necrosis factor containing a functional death domain. This association leads to the formation of the DISC, thereby inducing apoptosis. The entire process is initiated when the cell registers the presence of CD95L, the cognate ligand for APO-1. Upon binding, the CAP proteins and procaspase-8 (composed of FLICE, MACH, and Mch5) bind to CD95 through death domain and death effector domain interactions. Procaspase-8 activation is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death Effector Domain
The death-effector domain (DED) is a protein interaction domain found only in eukaryotes that regulates a variety of cellular signalling pathways. The DED domain is found in inactive procaspases (cysteine proteases) and proteins that regulate caspase activation in the apoptosis cascade such as FAS-associating death domain-containing protein (FADD). FADD recruits procaspase 8 and procaspase 10 into a death induced signaling complex (DISC). This recruitment is mediated by a homotypic interaction between the procaspase DED and a second DED that is death effector domain in an adaptor protein that is directly associated with activated TNF receptors. Complex formation allows proteolytic activation of procaspase into the active caspase form which results in the initiation of apoptosis (cell death). Structurally the DED domain are a subclass of protein motif known as the death fold and contains 6 alpha helices, that closely resemble the structure of the Death domain (DD). Structure DE ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death Receptor 5
Death receptor 5 (DR5), also known as TRAIL receptor 2 (TRAILR2) and tumor necrosis factor receptor superfamily member 10B (TNFRSF10B), is a cell surface receptor of the TNF-receptor superfamily that binds TRAIL and mediates apoptosis. Function The protein encoded by this gene is a member of the TNF-receptor superfamily, and contains an intracellular death domain. This receptor can be activated by tumor necrosis factor-related apoptosis inducing ligand (TNFSF10/TRAIL/APO-2L), and transduces apoptosis signal. Mice have a homologous gene, tnfrsf10b, that has been essential in the elucidation of the function of this gene in humans. Studies with FADD-deficient mice suggested that FADD, a death domain containing adaptor protein, is required for the apoptosis mediated by this protein. Interactions DR5 has been shown to interact with: * Caspase 8, * Caspase 10, * FADD, and * TRAIL. Cancer therapy Monoclonal antibodies targeting DR5 have been developed and are currently under c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Death Receptor 4
Death receptor 4 (DR4), also known as TRAIL receptor 1 (TRAILR1) and tumor necrosis factor receptor superfamily member 10A (TNFRSF10A), is a cell surface receptor of the TNF-receptor superfamily that binds TRAIL and mediates apoptosis. Function The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptor is activated by tumor necrosis factor-related apoptosis inducing ligand (TNFSF10/TRAIL), and thus transduces cell death signal and induces cell apoptosis. Studies with FADD-deficient mice suggested that FADD, a death domain containing adaptor protein, is required for the apoptosis mediated by this protein. Interactions TNFRSF10A has been shown to interact with DAP3 28S ribosomal protein S29, mitochondrial, also known as death-associated protein 3 (DAP3), is a protein that in humans is encoded by the ''DAP3'' gene on chromosome 1. This gene encodes a 28S subunit protein of the mitochondrial ribosome (mitori .... References Further reading ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRAIL
A trail, also known as a path or track, is an unpaved lane or small road usually passing through a natural area. In the United Kingdom and the Republic of Ireland, a path or footpath is the preferred term for a pedestrian or hiking trail. The term is also applied in North America to routes along rivers, and sometimes to highways. In the US, the term was historically used for a route into or through wild territory used by explorers and migrants (e.g. the Oregon Trail). In the United States, "trace" is a synonym for trail, as in Natchez Trace. Some trails are dedicated only for walking, cycling, horse riding, snowshoeing or cross-country skiing, but not more than one use; others, as in the case of a bridleway in the UK, are multi-use and can be used by walkers, cyclists and equestrians alike. There are also unpaved trails used by dirt bikes and other off-road vehicles, and in some places, like the Alps, trails are used for moving cattle and other livestock. Usage In Austra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-PKcs
DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as ''PRKDC'' or ''XRCC7''. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase comprising a single polypeptide chain of 4,128 amino acids. Function DNA-PKcs is the catalytic subunit of a nuclear DNA-dependent serine/threonine protein kinase called DNA-PK. The second component is the autoimmune antigen Ku. On its own, DNA-PKcs is inactive and relies on Ku to direct it to DNA ends and trigger its kinase activity. DNA-PKcs is required for the non-homologous end joining (NHEJ) pathway of DNA repair, which rejoins double-strand breaks. It is also required for V(D)J recombination, a process that utilizes NHEJ to promote immune system diversity. DNA-PKcs knockout mice have severe combined immunodeficiency due to their V(D)J recombination defect. Many proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CrmA
{{disambig ...
CRMA may refer to: * City and Regional Magazine Association * Chulachomklao Royal Military Academy * Civil Rights Movement Archive *Canadian Radio Music Awards The Canadian Radio Music Awards were an annual series of awards presented by the Canadian Association of Broadcasters that was part of Canadian Music Week. The award show ran from 2016 to 2018. There has been no information on the future of the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FLICE
Caspase-8 is a caspase protein, encoded by the ''CASP8'' gene. It most likely acts upon caspase-3. ''CASP8'' orthologs have been identified in numerous mammals for which complete genome data are available. These unique orthologs are also present in birds. Function The ''CASP8'' gene encodes a member of the cysteine- aspartic acid protease ( caspase) family. Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. Caspases exist as inactive proenzymes composed of a prodomain, a large protease subunit, and a small protease subunit. Activation of caspases requires proteolytic processing at conserved internal aspartic residues to generate a heterodimeric enzyme consisting of the large and small subunits. This protein is involved in the programmed cell death induced by Fas and various apoptotic stimuli. The N-terminal FADD-like death effector domain of this protein suggests that it may interact with Fas-interacting protein FADD. This p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine Protease
Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad. Discovered by Gopal Chunder Roy in 1873, the first cysteine protease to be isolated and characterized was papain, obtained from ''Carica papaya''. Cysteine proteases are commonly encountered in fruits including the papaya, pineapple, fig and kiwifruit. The proportion of protease tends to be higher when the fruit is unripe. In fact, the latex of dozens of different plant families are known to contain cysteine proteases. Cysteine proteases are used as an ingredient in meat tenderizers. Classification The MEROPS protease classification system counts 14 superfamilies plus several currently unassigned families (as of 2013) each containing many families. Each superfamily uses the catalytic triad or dyad in a different protein fold and so represent convergent evolutio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
A dimer () ('' di-'', "two" + ''-mer'', "parts") is an oligomer consisting of two monomers joined by bonds that can be either strong or weak, covalent or intermolecular. Dimers also have significant implications in polymer chemistry, inorganic chemistry, and biochemistry. The term ''homodimer'' is used when the two molecules are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Under special conditions, most OH-containing molecules form dimers, e.g. the water dimer. Excimers and exciplexes are excited structures with a short lifetime. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
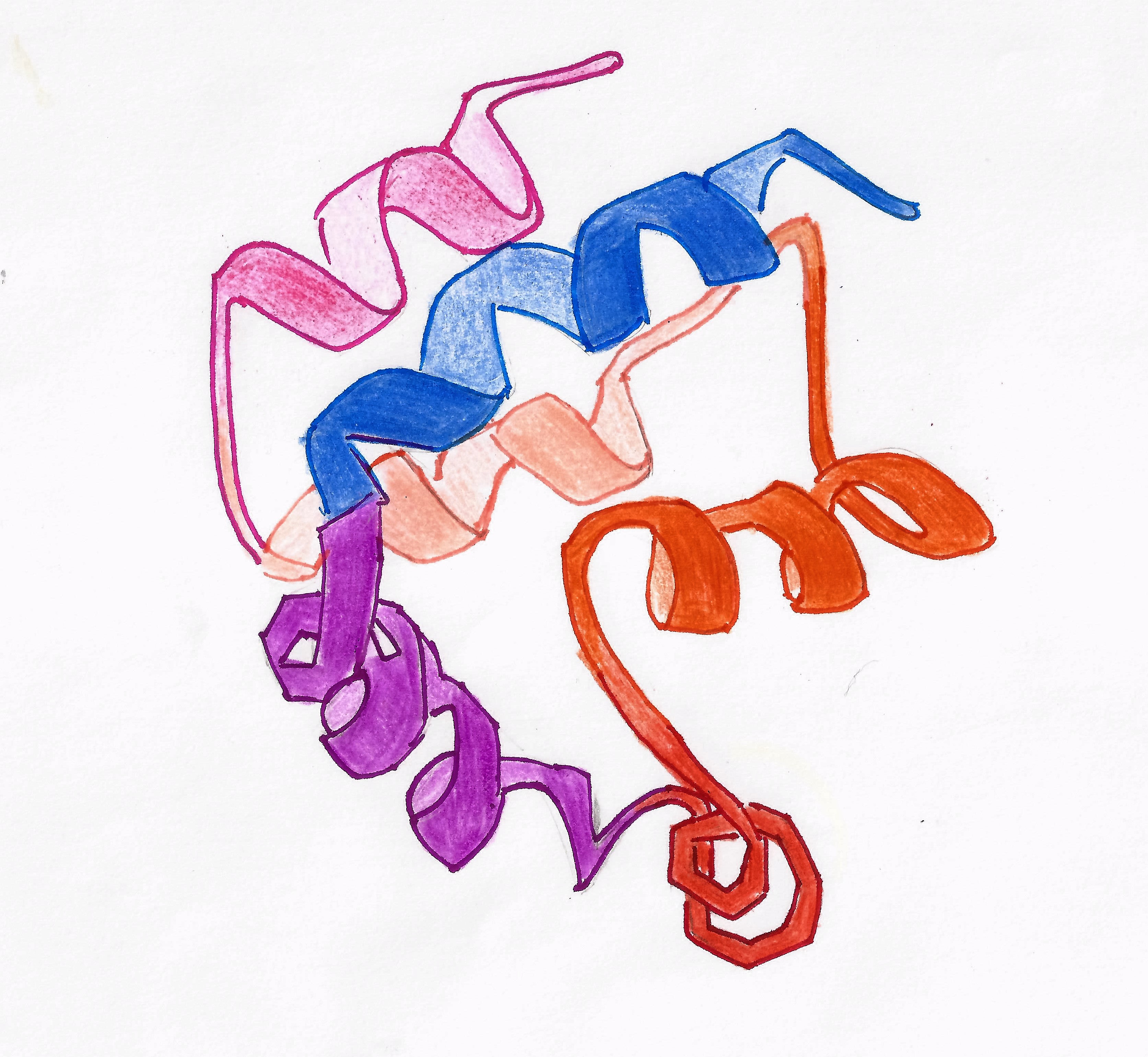
Death Domain
Death is the irreversible cessation of all biological functions that sustain an organism. For organisms with a brain, death can also be defined as the irreversible cessation of functioning of the whole brain, including brainstem, and brain death is sometimes used as a legal definition of death. The remains of a former organism normally begin to decompose shortly after death. Death is an inevitable process that eventually occurs in almost all organisms. Death is generally applied to whole organisms; the similar process seen in individual components of an organism, such as cells or tissues, is necrosis. Something that is not considered an organism, such as a virus, can be physically destroyed but is not said to die. As of the early 21st century, over 150,000 humans die each day, with ageing being by far the most common cause of death. Many cultures and religions have the idea of an afterlife, and also may hold the idea of judgement of good and bad deeds in one's life (heav ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)

