|
Drymoreomys
''Drymoreomys'' is a rodent genus in the tribe Oryzomyini that lives in the Atlantic Forest of Brazil. The single species, ''D. albimaculatus'', is known only from the states of São Paulo and Santa Catarina and was not named until 2011. It lives in the humid forest on the eastern slopes of the Serra do Mar and perhaps reproduces year-round. Although its range is relatively large and includes some protected areas, it is patchy and threatened, and the discoverers recommend that the animal be considered "Near Threatened" on the IUCN Red List. Within Oryzomyini, ''Drymoreomys'' appears to be most closely related to ''Eremoryzomys'' from the Andes of Peru, a biogeographically unusual relationship, in that the two populations are widely separated and each is adapted to an arid or a moist environment. With a body mass of , ''Drymoreomys'' is a medium-sized rodent with long fur that is orange to reddish-buff above and grayish with several white patches below. The pads on the hind ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eremoryzomys
''Eremoryzomys polius'', also known as the gray rice rat or the Marañon oryzomys,Musser and Carleton, 2005, p. 1153 is a rodent species in the tribe Oryzomyini of the family Cricetidae. Discovered in 1912 and first described in 1913 by Wilfred Osgood, it was originally placed in ''Oryzomys'' and named ''Oryzomys polius''. In 2006, a cladistics, cladistic analysis found that it was not closely related to ''Oryzomys'' in the strict sense or to any other oryzomyine then known, so that it is now placed in its own genus, ''Eremoryzomys''. The Brazilian genus ''Drymoreomys'', named in 2011, is probably the closest relative of ''Eremoryzomys''. ''Eremoryzomys'' has a limited distribution in the dry upper valley of the Marañón River in central Peru, but may yet contain more than one species. A large, long-tailed rice rat, with head and body length of , ''E. polius'' has gray fur and short ears. There are well-developed ungual tufts of hair on the hindfeet. Females have eight mammae ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eremoryzomys Polius
''Eremoryzomys polius'', also known as the gray rice rat or the Marañon oryzomys,Musser and Carleton, 2005, p. 1153 is a rodent species in the tribe Oryzomyini of the family Cricetidae. Discovered in 1912 and first described in 1913 by Wilfred Osgood, it was originally placed in ''Oryzomys'' and named ''Oryzomys polius''. In 2006, a cladistic analysis found that it was not closely related to ''Oryzomys'' in the strict sense or to any other oryzomyine then known, so that it is now placed in its own genus, ''Eremoryzomys''. The Brazilian genus ''Drymoreomys'', named in 2011, is probably the closest relative of ''Eremoryzomys''. ''Eremoryzomys'' has a limited distribution in the dry upper valley of the Marañón River in central Peru, but may yet contain more than one species. A large, long-tailed rice rat, with head and body length of , ''E. polius'' has gray fur and short ears. There are well-developed ungual tufts of hair on the hindfeet. Females have eight mammae. The rostr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oryzomyini
Oryzomyini is a tribe of rodents in the subfamily Sigmodontinae of the family Cricetidae. It includes about 120 species in about thirty genera,Weksler et al., 2006, table 1 distributed from the eastern United States to the southernmost parts of South America, including many offshore islands. It is part of the clade Oryzomyalia, which includes most of the South American Sigmodontinae. The name ''Oryzomyini'' derives from that of its type genus, '' Oryzomys'', which means "rice rat" or "rice mouse". Many species are also known as rice rats. Taxonomy Contents of Oryzomyini An oryzomyine group was first envisaged by Oldfield Thomas in the early 20th century. He defined it to include pentalophodont species, which have a mesoloph(id) on the upper and lower molars, with a long palate (extending past the third molars). Thomas included '' Oligoryzomys'', ''Oecomys'', and '' Oryzomys'' (which included many species now in other genera), as well as '' Rhagomys'', which is currently class ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alexandre Reis Percequillo
Alexandre may refer to: * Alexandre (given name) * Alexandre (surname) * Alexandre (film) See also * Alexander Alexander is a male given name. The most prominent bearer of the name is Alexander the Great, the king of the Ancient Greek kingdom of Macedonia who created one of the largest empires in ancient history. Variants listed here are Aleksandar, Al ... * Xano (other), a Portuguese hypocoristic of the name "Alexandre" {{Disambig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
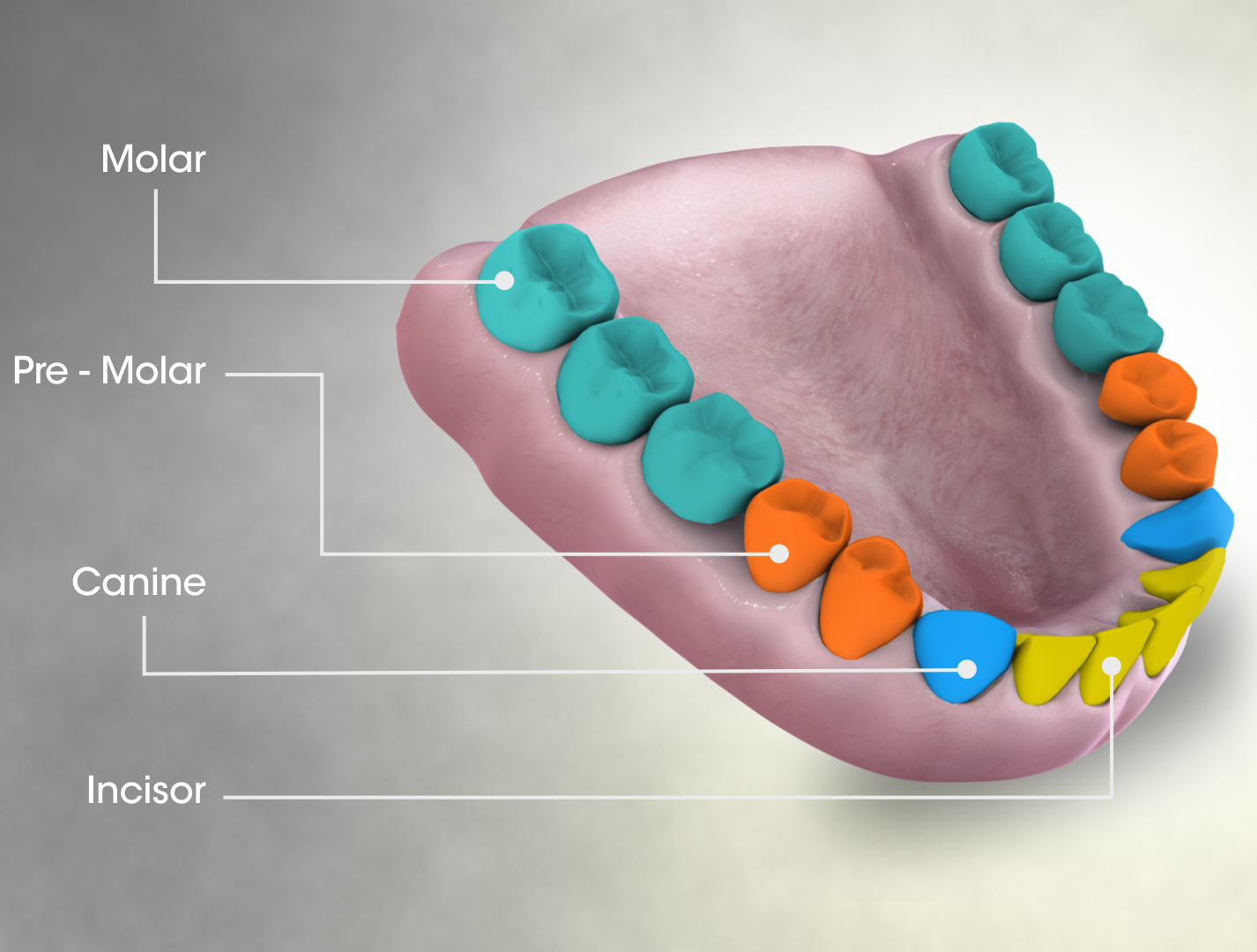
Molar (tooth)
The molars or molar teeth are large, flat teeth at the back of the mouth. They are more developed in mammals. They are used primarily to grind food during chewing. The name ''molar'' derives from Latin, ''molaris dens'', meaning "millstone tooth", from ''mola'', millstone and ''dens'', tooth. Molars show a great deal of diversity in size and shape across mammal groups. The third molar of humans is sometimes vestigial. Human anatomy In humans, the molar teeth have either four or five cusps. Adult humans have 12 molars, in four groups of three at the back of the mouth. The third, rearmost molar in each group is called a wisdom tooth. It is the last tooth to appear, breaking through the front of the gum at about the age of 20, although this varies from individual to individual. Race can also affect the age at which this occurs, with statistical variations between groups. In some cases, it may not even erupt at all. The human mouth contains upper (maxillary) and lower (mandib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Signal
Phylogenetic signal is an evolutionary and ecological term, that describes the tendency or the pattern of related biological species to resemble each other more than any other species that is randomly picked from the same phylogenetic tree. Characteristics Phylogenetic signal is usually described as the tendency of related biological species to resemble each other more than any other species that is randomly picked from the same phylogenetic tree. In other words, phylogenetic signal can be defined as the statistical dependence among species' trait values that is a consequence of their phylogenetic relationships. The traits (eg. morphological, ecological, life-history or behavioural traits) are inherited characteristics – meaning the trait values are usually alike within closely related species, while trait values of distantly related biological species do not resemble each other to a such great degree. It is often said that traits that are more similar in closely related tax ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saturation (genetic)
Genetic saturation is the result of multiple substitutions at the same site in a sequence, or identical substitutions in different sequence, such that the apparent sequence divergence rate is lower than the actual divergence that has occurred. In phylogenetics, saturation effects result in long branch attraction, where the most distant lineages have misleadingly short branch lengths. It also decreases phylogenetic information contained in the sequences. Genetic saturation occurs most rapidly on fast evolving sequences, such as the hypervariable region of mitochondrial DNA, or in Short tandem repeat such as on the Y-chromosome. See also *Long branch attraction *Molecular clock *Human mitochondrial molecular clock *Convergent evolution Convergent evolution is the independent evolution of similar features in species of different periods or epochs in time. Convergent evolution creates analogous structures that have similar form or function but were not present in the last co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clade
A clade (), also known as a monophyletic group or natural group, is a group of organisms that are monophyletic – that is, composed of a common ancestor and all its lineal descendants – on a phylogenetic tree. Rather than the English term, the equivalent Latin term ''cladus'' (plural ''cladi'') is often used in taxonomical literature. The common ancestor may be an individual, a population, or a species (extinct or extant). Clades are nested, one in another, as each branch in turn splits into smaller branches. These splits reflect evolutionary history as populations diverged and evolved independently. Clades are termed monophyletic (Greek: "one clan") groups. Over the last few decades, the cladistic approach has revolutionized biological classification and revealed surprising evolutionary relationships among organisms. Increasingly, taxonomists try to avoid naming taxa that are not clades; that is, taxa that are not monophyletic. Some of the relationships between org ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome B
Cytochrome b within both molecular and cell biology, is a protein found in the mitochondria of eukaryotic cells. It functions as part of the electron transport chain and is the main subunit of transmembrane cytochrome bc1 and b6f complexes. Function In the mitochondrion of eukaryotes and in aerobic prokaryotes, cytochrome b is a component of respiratory chain complex III () — also known as the bc1 complex or ubiquinol-cytochrome c reductase. In plant chloroplasts and cyanobacteria, there is an analogous protein, cytochrome b6, a component of the plastoquinone-plastocyanin reductase (), also known as the b6f complex. These complexes are involved in electron transport, the pumping of protons to create a proton-motive force ( PMF). This proton gradient is used for the generation of ATP. These complexes play a vital role in cells. Structure Cytochrome b/b6 is an integral membrane protein of approximately 400 amino acid residues that probably has 8 transmembrane segments. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial DNA
Mitochondrial DNA (mtDNA or mDNA) is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, such as adenosine triphosphate (ATP). Mitochondrial DNA is only a small portion of the DNA in a eukaryotic cell; most of the DNA can be found in the cell nucleus and, in plants and algae, also in plastids such as chloroplasts. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that the human mtDNA includes 16,569 base pairs and encodes 13 proteins. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It also permits an examination of the relatedness of populations, and so has become important in anthropology and biogeography. Origin Nuclear and mitochondrial DNA are thought to be of separate evolutionary origin, with the mtDNA being derive ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RBP3
Retinol-binding protein 3, interstitial (RBP3), also known as IRBP is a protein that in humans is encoded by the ''RBP3'' gene. RBP3 orthologs have been identified in most eutherians except tenrecs and armadillos. Function The inter- photoreceptor retinoid-binding protein is a large glycoprotein known to bind retinoids and found primarily in the interphotoreceptor matrix of the retina between the retinal pigment epithelium and the photoreceptor cells. It is thought to transport retinoids between the retinal pigment epithelium and the photoreceptors, a critical role in the visual process. Gene The human IRBP gene is approximately 9.5 kbp in length and consists of four exons separated by three introns. The introns are 1.6-1.9 kbp long. The gene is transcribed by photoreceptor and retinoblastoma cells into an approximately 4.3-kilobase mRNA that is translated and processed into a glycosylated protein of 135,000 Da. Structure The amino acid sequence of human IRBP can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear DNA
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. It adheres to Mendelian inheritance, with information coming from two parents, one male and one female—rather than matrilineally (through the mother) as in mitochondrial DNA. Structure Nuclear DNA is a nucleic acid, a polymeric biomolecule or biopolymer, found in the nucleus of eukaryotic cells. Its structure is a double helix, with two strands wound around each other, a structure first described by Francis Crick and James D. Watson (1953) using data collected by Rosalind Franklin. Each strand is a long polymer chain of repeating nucleotides. Each nucleotide is composed of a five-carbon sugar, a phosphate group, and an organic base. Nucleotides are distinguished by their bases: purines, large bases that include adenine and guanine; and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.png)
