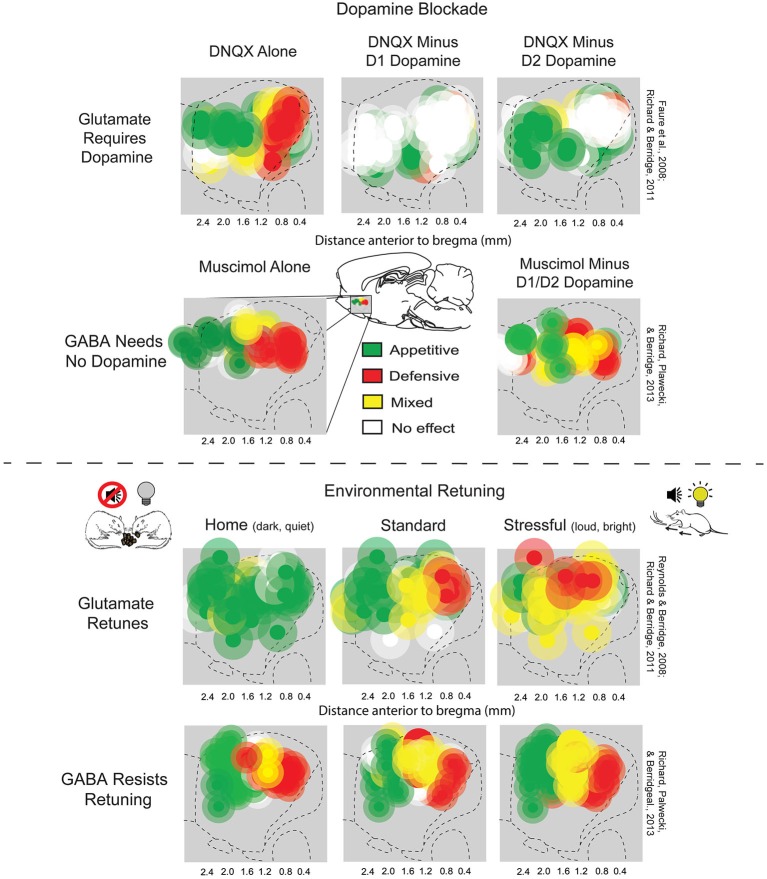
|
Dopamine Receptor D2
Dopamine receptor D2, also known as D2R, is a protein that, in humans, is encoded by the ''DRD2'' gene. After work from Paul Greengard's lab had suggested that dopamine receptors were the site of action of antipsychotic drugs, several groups, including those of Solomon Snyder and Philip Seeman used a radiolabeled antipsychotic drug to identify what is now known as the dopamine D2 receptor. The dopamine D2 receptor is the main receptor for most antipsychotic drugs. The structure of DRD2 in complex with the atypical antipsychotic risperidone has been determined. Function D2 receptors are coupled to Gi subtype of G protein. This G protein-coupled receptor inhibits adenylyl cyclase activity. In mice, regulation of D2R surface expression by the neuronal calcium sensor-1 (NCS-1) in the dentate gyrus is involved in exploration, synaptic plasticity and memory formation. Studies have shown potential roles for D2R in retrieval of fear memories in the prelimbic cortex and in discrimina ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleus Accumbens
The nucleus accumbens (NAc or NAcc; also known as the accumbens nucleus, or formerly as the ''nucleus accumbens septi'', Latin for "nucleus adjacent to the septum") is a region in the basal forebrain rostral to the preoptic area of the hypothalamus. The nucleus accumbens and the olfactory tubercle collectively form the ventral striatum. The ventral striatum and dorsal striatum collectively form the striatum, which is the main component of the basal ganglia. The dopaminergic neurons of the mesolimbic pathway project onto the GABAergic medium spiny neurons of the nucleus accumbens and olfactory tubercle. Each cerebral hemisphere has its own nucleus accumbens, which can be divided into two structures: the nucleus accumbens core and the nucleus accumbens shell. These substructures have different morphology and functions. Different NAcc subregions (core vs shell) and neuron subpopulations within each region (D1-type vs D2-type medium spiny neurons) are responsible for different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of ligan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Domain
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology Modeling
Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the "''target''" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the "''template''"). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been seen that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure. Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure. It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antagonist
An antagonist is a character in a story who is presented as the chief foe of the protagonist. Etymology The English word antagonist comes from the Greek ἀνταγωνιστής – ''antagonistēs'', "opponent, competitor, villain, enemy, rival," which is derived from ''anti-'' ("against") and ''agonizesthai'' ("to contend for a prize"). Types Heroes and villains The antagonist is commonly positioned against the protagonist and their world order. While most narratives will often portray the protagonist as a hero and the antagonist as a villain, like Harry Potter and Lord Voldemort in '' Harry Potter'', the antagonist does not always appear as the villain. In some narratives, like Light Yagami and L in '' Death Note'', the protagonist is a villain and the antagonist is an opposing hero. Antagonists are conventionally presented as making moral choices less savory than those of protagonists. This condition is often used by an author to create conflict within a story. This is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dopaminergic
Dopaminergic means "related to dopamine" (literally, "working on dopamine"), dopamine being a common neurotransmitter. Dopaminergic substances or actions increase dopamine-related activity in the brain. Dopaminergic brain pathways facilitate dopamine-related activity. For example, certain proteins such as the dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2), and dopamine receptors can be classified as dopaminergic, and neurons that synthesize or contain dopamine and synapses with dopamine receptors in them may also be labeled as ''dopaminergic''. Enzymes that regulate the biosynthesis or metabolism of dopamine such as aromatic L-amino acid decarboxylase or DOPA decarboxylase, monoamine oxidase (MAO), and catechol ''O''-methyl transferase (COMT) may be referred to as ''dopaminergic'' as well. Also, any endogenous or exogenous chemical substance that acts to affect dopamine receptors or dopamine release through indirect actions (for example, on neurons t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Agonist
An agonist is a chemical that activates a receptor to produce a biological response. Receptors are cellular proteins whose activation causes the cell to modify what it is currently doing. In contrast, an antagonist blocks the action of the agonist, while an inverse agonist causes an action opposite to that of the agonist. Etymology From the Greek αγωνιστής (agōnistēs), contestant; champion; rival < αγων (agōn), contest, combat; exertion, struggle < αγω (agō), I lead, lead towards, conduct; drive Types of agonists can be activated by either endogenous agonists (such as |
Synapse
In the nervous system, a synapse is a structure that permits a neuron (or nerve cell) to pass an electrical or chemical signal to another neuron or to the target effector cell. Synapses are essential to the transmission of nervous impulses from one neuron to another. Neurons are specialized to pass signals to individual target cells, and synapses are the means by which they do so. At a synapse, the plasma membrane of the signal-passing neuron (the ''presynaptic'' neuron) comes into close apposition with the membrane of the target (''postsynaptic'') cell. Both the presynaptic and postsynaptic sites contain extensive arrays of molecular machinery that link the two membranes together and carry out the signaling process. In many synapses, the presynaptic part is located on an axon and the postsynaptic part is located on a dendrite or soma. Astrocytes also exchange information with the synaptic neurons, responding to synaptic activity and, in turn, regulating neurotransmission. Syna ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.png)





