|
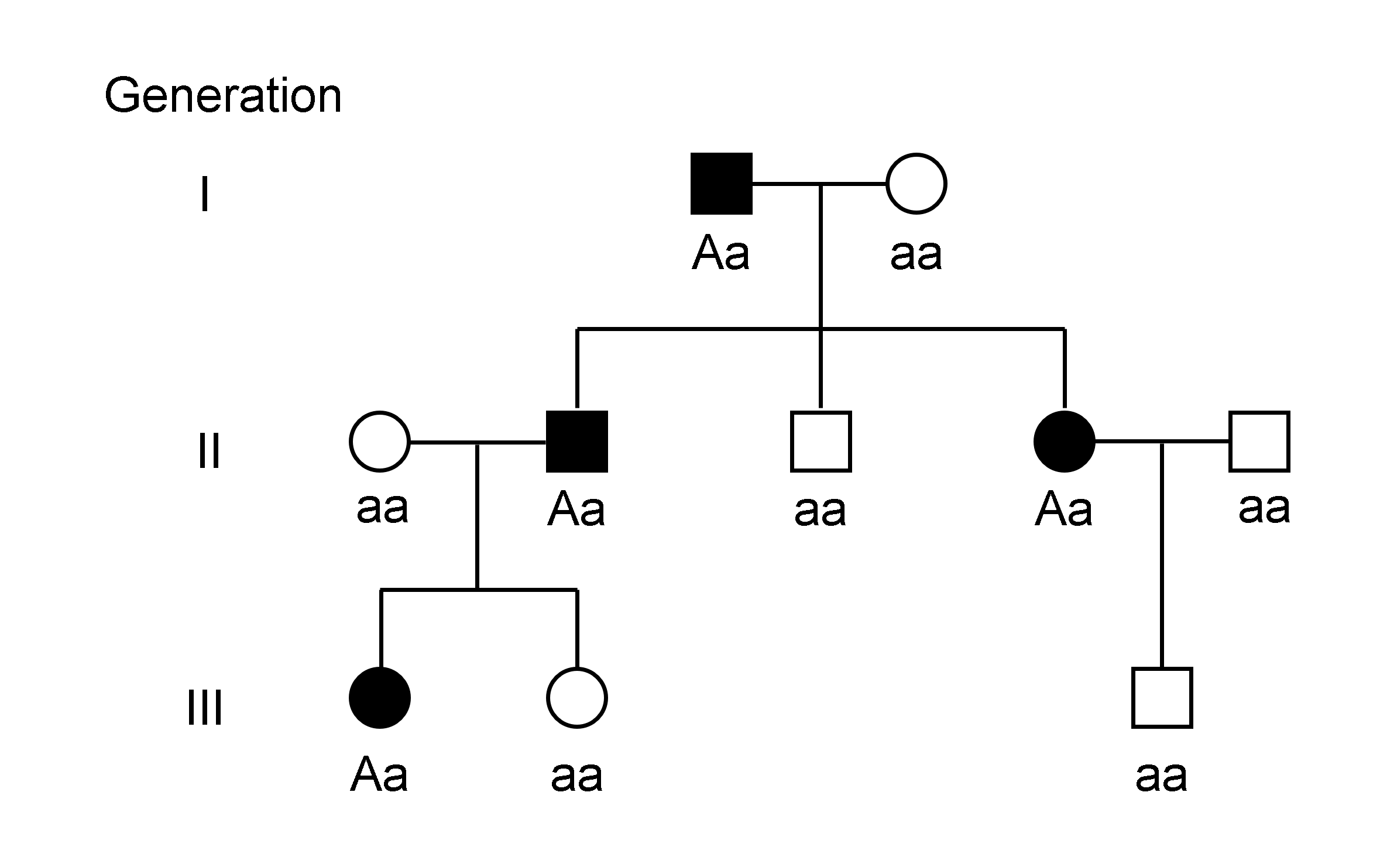
Disease Gene Identification
Disease gene identification is a process by which scientists identify the mutant genotypes responsible for an inherited genetic disorder. Mutations in these genes can include single nucleotide substitutions, single nucleotide additions/deletions, deletion of the entire gene, and other genetic abnormalities. Significance Knowledge of which genes (when non-functional) cause which disorders will simplify diagnosis of patients and provide insights into the functional characteristics of the mutation. The advent of modern-day high-throughput sequencing technologies combined with insights provided from the growing field of genomics is resulting in more rapid disease gene identification, thus allowing scientists to identify more complex mutations. Generic gene identification procedure Disease gene identification techniques often follow the same overall procedure. DNA is first collected from several patients who are believed to have the same genetic disease. Then, their DNA sam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plant. Howev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotype
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intronic
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRNA introns, group I introns, group II introns, and s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reference Genome
A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead a reference provides a haploid mosaic of different DNA sequences from each donor. For example, the most recent human reference genome (assembly '' GRCh38/hg38'') is derived from >60 genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several locations, using dedicated browsers s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Assembly
In bioinformatics, sequence assembly refers to aligning and merging fragments from a longer DNA sequence in order to reconstruct the original sequence. This is needed as DNA sequencing technology might not be able to 'read' whole genomes in one go, but rather reads small pieces of between 20 and 30,000 bases, depending on the technology used. Typically, the short fragments (reads) result from shotgun sequencing genomic DNA, or gene transcript ( ESTs). The problem of sequence assembly can be compared to taking many copies of a book, passing each of them through a shredder with a different cutter, and piecing the text of the book back together just by looking at the shredded pieces. Besides the obvious difficulty of this task, there are some extra practical issues: the original may have many repeated paragraphs, and some shreds may be modified during shredding to have typos. Excerpts from another book may also be added in, and some shreds may be completely unrecognizable. Genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exome Sequencing
Exome sequencing, also known as whole exome sequencing (WES), is a genomic technique for sequencing all of the protein-coding regions of genes in a genome (known as the exome). It consists of two steps: the first step is to select only the subset of DNA that encodes proteins. These regions are known as exons—humans have about 180,000 exons, constituting about 1% of the human genome, or approximately 30 million base pairs. The second step is to sequence the exonic DNA using any high-throughput DNA sequencing technology. The goal of this approach is to identify genetic variants that alter protein sequences, and to do this at a much lower cost than whole-genome sequencing. Since these variants can be responsible for both Mendelian and common polygenic diseases, such as Alzheimer's disease, whole exome sequencing has been applied both in academic research and as a clinical diagnostic. Motivation and comparison to other approaches Exome sequencing is especially effective in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tissue Culture
Tissue culture is the growth of tissues or cells in an artificial medium separate from the parent organism. This technique is also called micropropagation. This is typically facilitated via use of a liquid, semi-solid, or solid growth medium, such as broth or agar. Tissue culture commonly refers to the culture of animal cells and tissues, with the more specific term plant tissue culture being used for plants. The term "tissue culture" was coined by American pathologist Montrose Thomas Burrows. Historical use In 1885 Wilhelm Roux removed a section of the medullary plate of an embryonic chicken and maintained it in a warm saline solution for several days, establishing the basic principle of tissue culture. In 1907 the zoologist Ross Granville Harrison demonstrated the growth of frog embryonic cells that would give rise to nerve cells in a medium of clotted lymph. In 1913, E. Steinhardt, C. Israeli, and R. A. Lambert grew vaccinia virus in fragments of guinea pig corneal tiss ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotes
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: ''Bacteria'' (formerly Eubacteria) and ''Archaea'' (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes. Besides the absence of a nucleus, prokaryotes also lack mitochondria, or most of the other membrane-bound organelles that characterize the eukaryotic cell. It was once thought that prokaryotic cellular components within the cytopla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deletion Mapping
In genetics and especially genetic engineering, deletion mapping is a technique used to find out the mutation sites within a gene. The principle of deletion mapping involves crossing a strain which has a point mutation in a gene, with multiple strains who each carry a deletion in a different region of the same gene. Wherever recombination occurs between the two strains to produce a wild-type The wild type (WT) is the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard "normal" allele at a locus, in contrast to that produced by a non-standard, "m ... (+) gene (regardless of frequency), the point mutation cannot lie within the region of the deletion. If recombination cannot produce any wild-type genes, then it is reasonable to conclude that the point mutation and deletion are found within the same stretch of DNA. This example should demonstrate how the principle works: Suppose you have a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SiRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation. Text was copied from this source, which is available under Creative Commons Attribution 4.0 International License Structure Naturally occurring siRNAs have a well-defined structure that is a short (usually 20 to 24- bp) double-stranded RNA (dsRNA) with phosphorylated 5' ends and hydroxylated 3' ends with two overhanging nucleotides. The Dicer enzyme catalyzes production of siRNAs from long dsRNAs and small hairpin RNAs. siRNAs can also be introduced into cells by transfection. Since in principle any gene can be knocked down by a syntheti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Genetics
Reverse genetics is a method in molecular genetics that is used to help understand the function(s) of a gene by analysing the phenotypic effects caused by genetically engineering specific nucleic acid sequences within the gene. The process proceeds in the opposite direction to forward genetic screens of classical genetics. While forward genetics seeks to find the genetic basis of a phenotype or trait, reverse genetics seeks to find what phenotypes are controlled by particular genetic sequences. Automated DNA sequencing generates large volumes of genomic sequence data relatively rapidly. Many genetic sequences are discovered in advance of other, less easily obtained, biological information. Reverse genetics attempts to connect a given genetic sequence with specific effects on the organism. Reverse genetics systems can also allow the recovery and generation of infectious or defective viruses with desired mutations. This allows the ability to study the virus ''in vitro'' and ''in v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm '' Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |