|
DNA Electrophoresis
Nucleic acid electrophoresis is an analytical technique used to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules which are to be analyzed are set upon a viscous medium, the gel, where an electric field induces the nucleic acids (which are negatively charged due to their sugar-phosphate backbone) to migrate toward the anode (which is positively charged because this is an electrolytic rather than galvanic cell). The separation of these fragments is accomplished by exploiting the mobilities with which different sized molecules are able to pass through the gel. Longer molecules migrate more slowly because they experience more resistance within the gel. Because the size of the molecule affects its mobility, smaller fragments end up nearer to the anode than longer ones in a given period. After some time, the voltage is removed and the fragmentation gradient is analyzed. For larger separations between similar sized fragments, either the voltage or run time can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alkaline Gel Electrophoresis
In chemistry, an alkali (; from ar, القلوي, al-qaly, lit=ashes of the saltwort) is a basic, ionic salt of an alkali metal or an alkaline earth metal. An alkali can also be defined as a base that dissolves in water. A solution of a soluble base has a pH greater than 7.0. The adjective alkaline, and less often, alkalescent, is commonly used in English as a synonym for basic, especially for bases soluble in water. This broad use of the term is likely to have come about because alkalis were the first bases known to obey the Arrhenius definition of a base, and they are still among the most common bases. Etymology The word "alkali" is derived from Arabic ''al qalīy'' (or ''alkali''), meaning ''the calcined ashes'' (see calcination), referring to the original source of alkaline substances. A water-extract of burned plant ashes, called potash and composed mostly of potassium carbonate, was mildly basic. After heating this substance with calcium hydroxide (''slaked lime''), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Digest
A restriction digest is a procedure used in molecular biology to prepare DNA for analysis or other processing. It is sometimes termed ''DNA fragmentation'' (this term is used for other procedures as well). Hartl and Jones describe it this way: This enzymatic technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence at a particular location have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present.Hartl, Daniel L., Jones, Elizabeth W. (2001), ''Genetics: Analysis of Genes and Genomes'', Fifth Edition. The res ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Weight Size Marker
A molecular-weight size marker, also referred to as a protein ladder, DNA ladder, or RNA ladder, is a set of standards that are used to identify the approximate size of a molecule run on a gel during electrophoresis, using the principle that molecular weight is inversely proportional to migration rate through a gel matrix. Therefore, when used in gel electrophoresis, markers effectively provide a logarithmic scale by which to estimate the size of the other fragments (providing the fragment sizes of the marker are known). Protein, DNA, and RNA markers with pre-determined fragment sizes and concentrations are commercially available. These can be run in either agarose or polyacrylamide gels. The markers are loaded in lanes adjacent to sample lanes before the commencement of the run. DNA markers Development Although the concept of molecular-weight markers has been retained, techniques of development have varied throughout the years. New inventions of molecular-weight markers ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Supercoiled DNA
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twist s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twist s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Structure
A chemical structure determination includes a chemist's specifying the molecular geometry and, when feasible and necessary, the electronic structure of the target molecule or other solid. Molecular geometry refers to the spatial arrangement of atoms in a molecule and the chemical bonds that hold the atoms together, and can be represented using structural formulae and by molecular models; complete electronic structure descriptions include specifying the occupation of a molecule's molecular orbitals. Structure determination can be applied to a range of targets from very simple molecules (e.g., diatomic oxygen or nitrogen), to very complex ones (e.g., such as protein or DNA). Background Theories of chemical structure were first developed by August Kekulé, Archibald Scott Couper, and Aleksandr Butlerov, among others, from about 1858. These theories were first to state that chemical compounds are not a random cluster of atoms and functional groups, but rather had a definite order ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid Miniprep
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the inter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pulse Field Gel Electrophoresis
Pulsed field gel electrophoresis is a technique used for the separation of large DNA molecules by applying to a gel matrix an electric field that periodically changes direction. Historical background Standard gel electrophoresis techniques for separation of DNA molecules provided huge advantages for molecular biology research. However, it was unable to separate very large molecules of DNA effectively. DNA molecules larger than 15–20 kb migrating through a gel will essentially move together in a size-independent manner. At Columbia University in 1984, David C. Schwartz and Charles Cantor developed a variation on the standard protocol by introducing an alternating voltage gradient to improve the resolution of larger molecules. This technique became known as pulsed-field gel electrophoresis (PFGE). The development of PFGE expanded the range of resolution for DNA fragments by as much as two orders of magnitude. Procedure The procedure for this technique is relatively similar to p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Logarithm
In mathematics, the logarithm is the inverse function to exponentiation. That means the logarithm of a number to the base is the exponent to which must be raised, to produce . For example, since , the ''logarithm base'' 10 of is , or . The logarithm of to ''base'' is denoted as , or without parentheses, , or even without the explicit base, , when no confusion is possible, or when the base does not matter such as in big O notation. The logarithm base is called the decimal or common logarithm and is commonly used in science and engineering. The natural logarithm has the number as its base; its use is widespread in mathematics and physics, because of its very simple derivative. The binary logarithm uses base and is frequently used in computer science. Logarithms were introduced by John Napier in 1614 as a means of simplifying calculations. They were rapidly adopted by navigators, scientists, engineers, surveyors and others to perform high-a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intercalation (biochemistry)
In biochemistry, intercalation is the insertion of molecules between the planar bases of deoxyribonucleic acid (DNA). This process is used as a method for analyzing DNA and it is also the basis of certain kinds of poisoning. There are several ways molecules (in this case, also known as ''ligands'') can interact with DNA. Ligands may interact with DNA by covalently binding, electrostatically binding, or intercalating. Intercalation occurs when ligands of an appropriate size and chemical nature fit themselves in between base pairs of DNA. These ligands are mostly polycyclic, aromatic, and planar, and therefore often make good nucleic acid stains. Intensively studied DNA intercalators include berberine, ethidium bromide, proflavine, daunomycin, doxorubicin, and thalidomide. DNA intercalators are used in chemotherapeutic treatment to inhibit DNA replication in rapidly growing cancer cells. Examples include doxorubicin (adriamycin) and daunorubicin (both of which are used in treat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
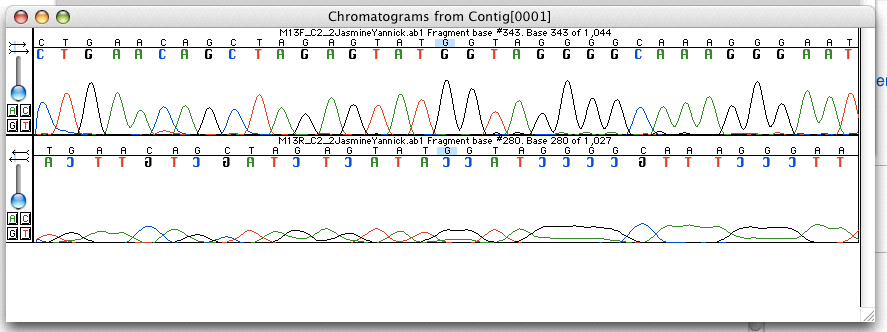
Electropherogram
An electropherogram, or electrophoregram, can also be referred to as an EPG or e-gram. It is a record or chart produced when electrophoresis is used in an analytical technique, primarily in the fields of forensic biology, molecular biology and biochemistry. The method utilizes data points that correspond with a specific time and fluorescence intensity at various wavelengths of light to represent a DNA profile. In the field of genetics, an electropherogram is a plot of DNA fragment sizes, typically used for genotyping such as DNA sequencing. The data is plotted with time, shown via base pairs (bps), on the x-axis and fluorescence intensity on the y-axis. Such plots are often achieved using an instrument such as an automated DNA sequencer paired with capillary electrophoresis (CE). Such electropherograms may be used to determine DNA sequence genotypes, or genotypes that are based on the length of specific DNA fragments or number of short tandem repeats (STR) at a specific locus b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |