|
Computational Genes
A computational gene is a molecular automaton consisting of a structural part and a functional part; and its design is such that it might work in a cellular environment. The structural part is a naturally occurring gene, which is used as a skeleton to encode the input and the transitions of the automaton (Fig. 1A). The conserved features of a structural gene (e.g., DNA polymerase binding site, start and stop codons, and splicing sites) serve as constants of the computational gene, while the coding regions, the number of exons and introns, the position of start and stop codon, and the automata theoretical variables (symbols, states, and transitions) are the design parameters of the computational gene. The constants and the design parameters are linked by several logical and biochemical constraints (e.g., encoded automata theoretic variables must not be recognized as splicing junctions). The input of the automaton are molecular markers given by single stranded DNA (ssDNA) molecules. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of Outline of life forms, life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal Kingdom (biology), kingdom Asgard (Archaea), Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only Two-domain system, two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as Flagellated cell, flagellated cells. The leading evolutiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
DNA Computing
DNA computing is an emerging branch of unconventional computing which uses DNA, biochemistry, and molecular biology hardware, instead of the traditional electronic computing. Research and development in this area concerns theory, experiments, and applications of DNA computing. Although the field originally started with the demonstration of a computing application by Len Adleman in 1994, it has now been expanded to several other avenues such as the development of storage technologies, nanoscale imaging modalities, synthetic controllers and reaction networks, etc. History Leonard Adleman of the University of Southern California initially developed this field in 1994. — The first DNA computing paper. Describes a solution for the directed Hamiltonian path problem. Also available here: Adleman demonstrated a proof-of-concept use of DNA as a form of computation which solved the seven-point Hamiltonian path problem. Since the initial Adleman experiments, advances have occur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Cyclic Enzyme System
A cyclic enzyme system is a theoretical system of two enzymes sharing a single substrate or cofactor, also referred to as a biochemical switching device. It has been used as a biochemical implementation of a simple computational device, acting as a chemical diode. See also * Biocomputer * Computational gene A computational gene is a molecular automaton consisting of a structural part and a functional part; and its design is such that it might work in a cellular environment. The structural part is a naturally occurring gene, which is used as a skelet ... References Enzymes {{biotech-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Biocomputers
Biological computers use biologically derived molecules — such as DNA and/or proteins — to perform digital or real computations. The development of biocomputers has been made possible by the expanding new science of nanobiotechnology. The term nanobiotechnology can be defined in multiple ways; in a more general sense, nanobiotechnology can be defined as any type of technology that uses both nano-scale materials (i.e. materials having characteristic dimensions of 1-100 nanometers) and biologically based materials. A more restrictive definition views nanobiotechnology more specifically as the design and engineering of proteins that can then be assembled into larger, functional structures The implementation of nanobiotechnology, as defined in this narrower sense, provides scientists with the ability to engineer biomolecular systems specifically so that they interact in a fashion that can ultimately result in the computational functionality of a computer. Scientific background B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Sense (molecular Biology)
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, the negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript. DNA sense Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Nucleases
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning. There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the ''middle'' of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. History In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichia col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
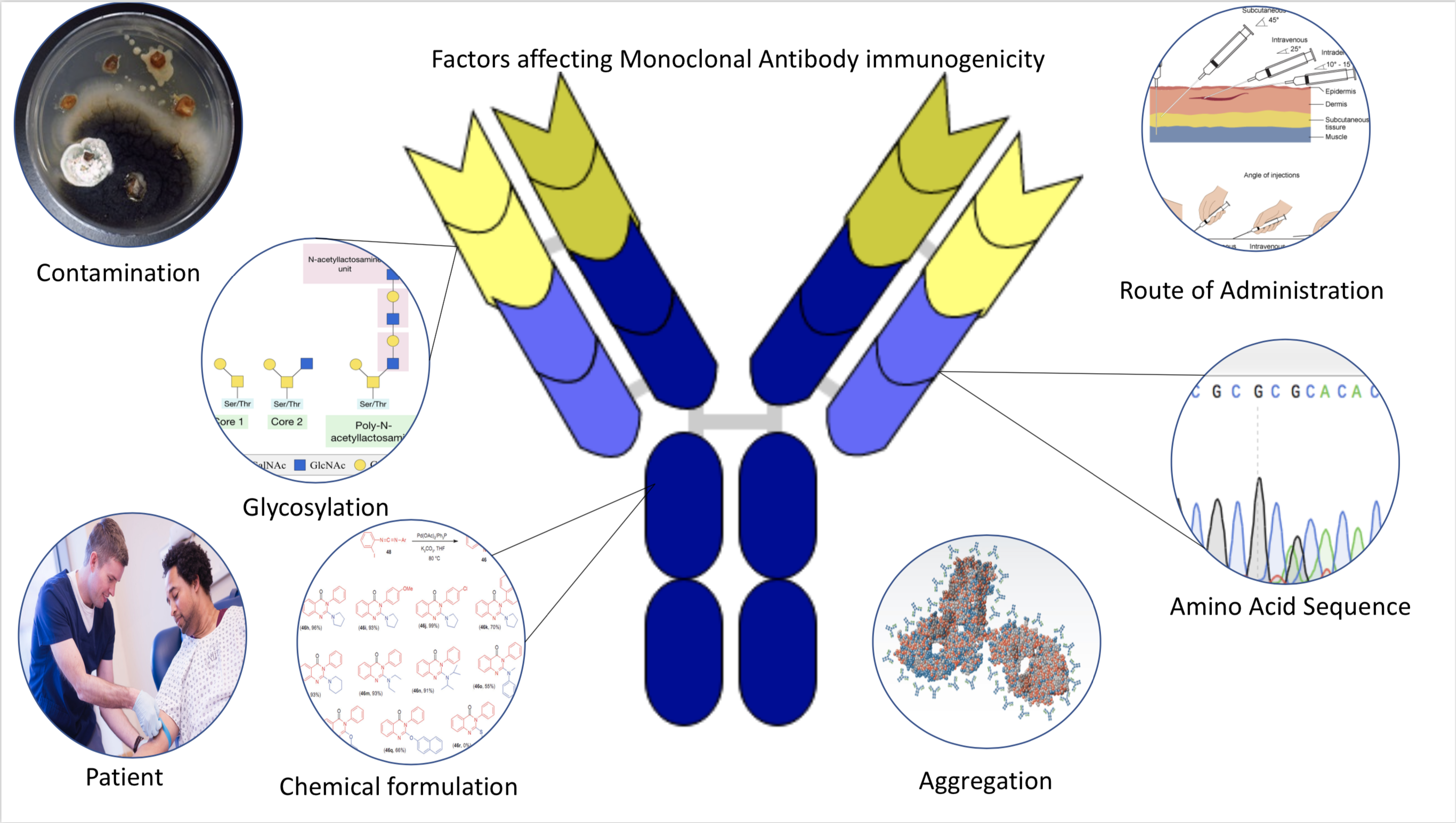
Immunogenicity
Immunogenicity is the ability of a foreign substance, such as an antigen, to provoke an immune response in the body of a human or other animal. It may be wanted or unwanted: * Wanted immunogenicity typically relates to vaccines, where the injection of an antigen (the vaccine) provokes an immune response against the pathogen, protecting the organism from future exposure. Immunogenicity is a central aspect of vaccine development. * Unwanted immunogenicity is an immune response by an organism against a therapeutic antigen. This reaction leads to production of anti-drug-antibodies (ADAs), inactivating the therapeutic effects of the treatment and potentially inducing adverse effects. A challenge in biotherapy is predicting the immunogenic potential of novel protein therapeutics. For example, immunogenicity data from high-income countries are not always transferable to low-income and middle-income countries. Another challenge is considering how the immunogenicity of vaccines changes wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Drug Delivery
Drug delivery involves various methods and technologies designed to transport pharmaceutical compounds to their target sites helping therapeutic effect. It involves principles related to drug preparation, route of administration, site-specific targeting, metabolism, and toxicity all aimed to optimize efficacy and safety, while improving patient convenience and compliance. A key goal of drug delivery is to modify a drug's pharmacokinetics and specificity by combining it with different excipients, drug carriers, and medical devices designed to control its distribution and activity in the body. Enhancing bioavailability and prolonging duration of action are essential strategies for improving therapeutic outcomes, particularly in chronic disease management. Additionally, some research emphasizes on improving safety for the individuals administering the medication. For example, microneedle patches have been developed for vaccines and drug delivery to minimize the risk of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Biological Membrane
A biological membrane, biomembrane or cell membrane is a selectively permeable membrane that separates the interior of a cell from the external environment or creates intracellular compartments by serving as a boundary between one part of the cell and another. Biological membranes, in the form of eukaryotic cell membranes, consist of a phospholipid bilayer with embedded, integral and peripheral proteins used in communication and transportation of chemicals and ions. The bulk of lipids in a cell membrane provides a fluid matrix for proteins to rotate and laterally diffuse for physiological functioning. Proteins are adapted to high membrane fluidity environment of the lipid bilayer with the presence of an annular lipid shell, consisting of lipid molecules bound tightly to the surface of integral membrane proteins. The cell membranes are different from the isolating tissues formed by layers of cells, such as mucous membranes, basement membranes, and serous membranes. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Cell Nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, no nuclei, and a few others including osteoclasts have Multinucleate, many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long strands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are Nuclear organization, structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |