|
Cancer Epigenetics
Cancer epigenetics is the study of epigenetics, epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than mutation, genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of gene expression, expression of genes that occurs about 10 times more frequently by transcription silencing (caused by epigenetic promoter hypermethylation of CpG site#Methylation, silencing, cancer, and aging, CpG islands) than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 Genetic hitchhiking, hitchhiker or passenger mutations. However, in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repeat Sequences
Repeated sequences (also known as repetitive elements, repeating units or repeats) are short or long patterns of nucleic acids (DNA or RNA) that occur in multiple copies throughout the genome. In many organisms, a significant fraction of the genomic DNA is repetitive, with over two-thirds of the sequence consisting of repetitive elements in humans. Some of these repeated sequences are necessary for maintaining important genome structures such as telomeres or centromeres. Repeated sequences are categorized into different classes depending on features such as structure, length, location, origin, and mode of multiplication. The disposition of repetitive elements throughout the genome can consist either in directly-adjacent arrays called tandem repeats or in repeats dispersed throughout the genome called interspersed repeats. Tandem repeats and interspersed repeats are further categorized into subclasses based on the length of the repeated sequence and/or the mode of multiplication. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase. There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency. Any piece of DNA with the point centromere DNA sequence on it will typically form a centromere if pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Noncoding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regulatory RNAs). Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some non-coding regions appear to be mostly nonfunctional such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. Fraction of non-coding genomic DNA In bacteria, the coding regions typically take up 88 % of the genome. The remaining 12 % consists largely of non-coding genes and regulatory sequences, which means that almost all of the bacterial genome has a function. The amount of coding DNA in eukaryrotes is usually a much smaller fraction of the genome because eukaryotic genomes contai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Colorectal Cancer
Colorectal cancer (CRC), also known as bowel cancer, colon cancer, or rectal cancer, is the development of cancer from the colon or rectum (parts of the large intestine). Signs and symptoms may include blood in the stool, a change in bowel movements, weight loss, and fatigue. Most colorectal cancers are due to old age and lifestyle factors, with only a small number of cases due to underlying genetic disorders. Risk factors include diet, obesity, smoking, and lack of physical activity. Dietary factors that increase the risk include red meat, processed meat, and alcohol. Another risk factor is inflammatory bowel disease, which includes Crohn's disease and ulcerative colitis. Some of the inherited genetic disorders that can cause colorectal cancer include familial adenomatous polyposis and hereditary non-polyposis colon cancer; however, these represent less than 5% of cases. It typically starts as a benign tumor, often in the form of a polyp, which over time becomes cancerous. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Insulin-like Growth Factor
The insulin-like growth factors (IGFs) are proteins with high sequence similarity to insulin. IGFs are part of a complex system that cells use to communicate with their physiologic environment. This complex system (often referred to as the IGF "axis") consists of two cell-surface receptors (IGF1R and IGF2R), two ligands (IGF-1 and IGF-2), a family of seven high-affinity IGF-binding proteins (IGFBP1 to IGFBP7), as well as associated IGFBP degrading enzymes, referred to collectively as proteases. IGF1/GH axis The IGF "axis" is also commonly referred to as the Growth Hormone/IGF-1 Axis. Insulin-like growth factor 1 (commonly referred to as IGF-1 or at times using Roman numerals as IGF-I) is mainly secreted by the liver as a result of stimulation by growth hormone (GH). IGF-1 is important for both the regulation of normal physiology, as well as a number of pathological states, including cancer. The IGF axis has been shown to play roles in the promotion of cell proliferation and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposable Element
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transposition often results in duplication of the same genetic material. Barbara McClintock's discovery of them earned her a Nobel Prize in 1983. Its importance in personalized medicine is becoming increasingly relevant, as well as gaining more attention in data analytics given the difficulty of analysis in very high dimensional spaces. Transposable elements make up a large fraction of the genome and are responsible for much of the mass of DNA in a eukaryotic cell. Although TEs are selfish genetic elements, many are important in genome function and evolution. Transposons are also very useful to researchers as a means to alter DNA inside a living organism. There are at least two classes of TEs: Class I TEs or retrotransposons generally functi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome Instability
Chromosomal instability (CIN) is a type of genomic instability in which chromosomes are unstable, such that either whole chromosomes or parts of chromosomes are duplicated or deleted. More specifically, CIN refers to the increase in rate of addition or loss of entire chromosomes or sections of them. The unequal distribution of DNA to daughter cells upon mitosis results in a failure to maintain euploidy (the correct number of chromosomes) leading to aneuploidy (incorrect number of chromosomes). In other words, the daughter cells do not have the same number of chromosomes as the cell they originated from. Chromosomal instability is the most common form of genetic instability and cause of aneuploidy. These changes have been studied in solid tumors (a tumor that usually doesn't contain liquid, pus, or air, compared to liquid tumor), which may or may not be cancerous. CIN is a common occurrence in solid and haematological cancers, especially colorectal cancer. Although many tumours sho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
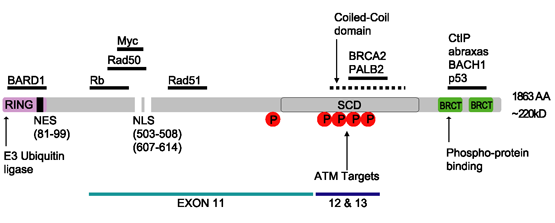
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and '' BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissue, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a normal, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 have also been studied in other organisms including mouse and the budding yeast ''Saccharomyces cerevisiae''. Function This gene was identified as a locus frequently mutated in hereditary nonpolyposis colon cancer. It is a human homolog of the ''E. coli'' DNA mismatch repair gene, mutL, which mediates protein-protein interactions during mismatch recognition, strand discrimination, and strand removal. Defects in MLH1 are associated with the microsatellite instability observed in hereditary nonpolyposis colon cancer. Alternatively spliced transcript variants encoding different isoforms have been described, but their full-length natures have not been determined. Role in DNA mismatch repair MLH1 protein is one component of a system of seve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenomatous Polyposis Coli
Adenomatous polyposis coli (APC) also known as deleted in polyposis 2.5 (DP2.5) is a protein that in humans is encoded by the ''APC'' gene. The APC protein is a negative regulator that controls beta-catenin concentrations and interacts with E-cadherin, which are involved in cell adhesion. Mutations in the ''APC'' gene may result in colorectal cancer. ''APC'' is classified as a tumor suppressor gene. Tumor suppressor genes prevent the uncontrolled growth of cells that may result in cancerous tumors. The protein made by the ''APC'' gene plays a critical role in several cellular processes that determine whether a cell may develop into a tumor. The APC protein helps control how often a cell divides, how it attaches to other cells within a tissue, how the cell polarizes and the morphogenesis of the 3D structures, or whether a cell moves within or away from tissue. This protein also helps ensure that the chromosome number in cells produced through cell division is correct. The APC pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |