|
Cytochrome P450 Cam
In enzymology, a camphor 5-monooxygenase () is an enzyme that catalyzes the chemical reaction :(+)-camphor + putidaredoxin + O2 \rightleftharpoons (+)-exo-5-hydroxycamphor + oxidized putidaredoxin + H2O The 3 substrates of this enzyme are (+)-camphor, putidaredoxin, and O2, whereas its 3 products are (+)-exo-5-hydroxycamphor, oxidized putidaredoxin, and H2O. This enzyme belongs to the family of oxidoreductases, specifically those acting on paired donors, with O2 as oxidant and incorporation or reduction of oxygen. The oxygen incorporated need not be derived from O2 with reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen into the other donor. The systematic name of this enzyme class is (+)-camphor,reduced putidaredoxin:oxygen oxidoreductase (5-hydroxylating). Other names in common use include camphor 5-exo-methylene hydroxylase, 2-bornanone 5-exo-hydroxylase, bornanone 5-exo-hydroxylase, camphor 5-exo-hydroxylase, camphor 5-exohydroxylase, camp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudomonas Putida
''Pseudomonas putida'' is a Gram-negative, rod-shaped, saprotrophic soil bacterium. Based on 16S rRNA analysis, ''P. putida'' was taxonomically confirmed to be a ''Pseudomonas'' species (''sensu stricto'') and placed, along with several other species, in the ''P. putida'' group, to which it lends its name. However, phylogenomic analysis of complete genomes from the entire ''Pseudomonas'' genus clearly showed that the genomes that were named as ''P. putida'' did not form a monophyletic clade, but were dispersed and formed a wider evolutionary group (the putida group) that included other species as well, such as ''P. alkylphenolia, P. alloputida, P. monteilii, P. cremoricolorata, P. fulva, P. parafulva, P. entomophila, P. mosselii, P. plecoglossicida'' and several genomic species (new species which are currentely not validely defined as new species. A variety of ''P. putida'', called multiplasmid hydrocarbon-degrading ''Pseudomonas'', is the first patented organism in th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADH
Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD and NADH (H for hydrogen), respectively. In metabolism, nicotinamide adenine dinucleotide is involved in redox reactions, carrying electrons from one reaction to another. The cofactor is, therefore, found in two forms in cells: NAD is an oxidizing agent – it accepts electrons from other molecules and becomes reduced. This reaction, also with H+, forms NADH, which can then be used as a reducing agent to donate electrons. These electron transfer reactions are the main function of NAD. However, it is also used in other cellular processes, most notably as a substrate of enzymes in adding or removing chemical groups t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron–sulfur Protein
Iron–sulfur proteins (or iron–sulphur proteins in British spelling) are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulner ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome P450
Cytochromes P450 (CYPs) are a Protein superfamily, superfamily of enzymes containing heme as a cofactor (biochemistry), cofactor that functions as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important for the clearance (pharmacology), clearance of various compounds, as well as for hormone synthesis and breakdown. In 1963, Ronald W. Estabrook, Estabrook, David Y. Cooper, Cooper, and Otto Rosenthal, Rosenthal described the role of CYP as a catalyst in steroid hormone synthesis and drug metabolism. In plants, these proteins are important for the biosynthesis of secondary metabolite, defensive compounds, fatty acids, and hormones. CYP enzymes have been identified in all kingdom (biology), kingdoms of life: animals, plants, fungus, fungi, protists, bacteria, and archaea, as well as in viruses. However, they are not omnipresent; for example, they have not been found in ''Escherichia coli''. , more than 300,000 distinct CYP proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
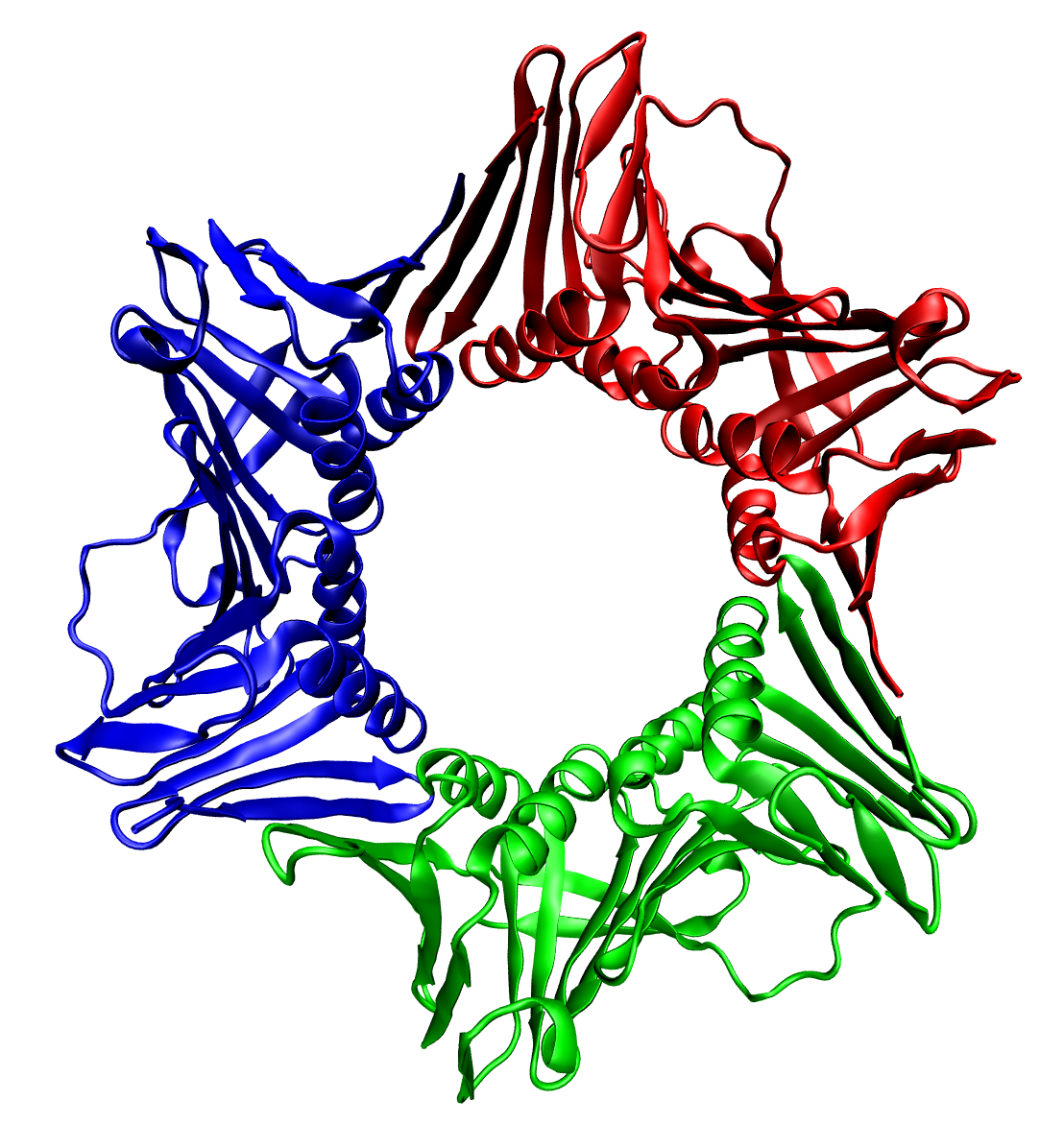
Protein Trimer
In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein. Porins usually arrange themselves in membranes as trimers. Bacteriophage T4 tail fiber Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation. The distal portion of each of the bacteriophage T4 tail fibers is encoded by ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information. Since many materials can form crystals—such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules—X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among various mat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Camphor
Camphor () is a waxy, colorless solid with a strong aroma. It is classified as a terpenoid and a cyclic ketone. It is found in the wood of the camphor laurel ('' Cinnamomum camphora''), a large evergreen tree found in East Asia; and in the kapur tree ( ''Dryobalanops'' sp.), a tall timber tree from South East Asia. It also occurs in some other related trees in the laurel family, notably '' Ocotea usambarensis''. Rosemary leaves (''Rosmarinus officinalis'') contain 0.05 to 0.5% camphor, while camphorweed (''Heterotheca'') contains some 5%. A major source of camphor in Asia is camphor basil (the parent of African blue basil). Camphor can also be synthetically produced from oil of turpentine. The compound is chiral, existing in two possible enantiomers as shown in the structural diagrams. The structure on the left is the naturally occurring (+)-camphor ((1''R'',4''R'')-bornan-2-one), while its mirror image shown on the right is the (−)-camphor ((1''S'',4''S'')-bornan-2-one). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |